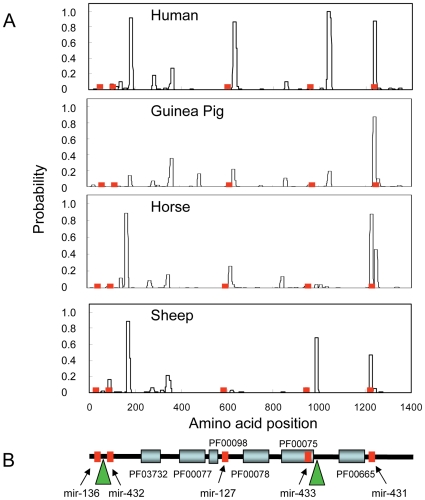
Figure 7. Structural domains in the PEG11 protein.
(A) Probability of coiled coil formation. All sequences were extracted from the UCSC genome browser (http://genome.ucsc.edu/). Coiled coils were predicted using Coils and a 14 amino acid window [52]. The relative positions in the PEG11 protein corresponding with regions encoding antisense miRNA are shown in red. (B) Structural domains common to mammalian PEG11 proteins. The diagram is based on the ovine sequence but is representative of mammalian sequences. Structural domains were identified by searching the Pfam protein families database [51]. PF03732 (capsid-like or retrotransposon gag protein domain; Expect score = 3.4e−11); PF00077 (retroviral aspartyl protease domain; Expect score = 0.33); PF00098 (zinc finger knuckle; Expect score not significant); PF00078 (reverse transcriptase domain; Expect score = 1.4e−5); PF00075 (RNaseH; Expect score not significant) domain; PF00665 (integrase core domain; Expect score not significant). Mutations to key residues in some of the PEG11 retrotransposon domains have compromised the significance of identification of some Pfam families, which are often biased in their weightings of catalytic residues. The positions corresponding to regions encoding antisense miRNA are shown in red. The green triangles denote positions where there are large insertions of repetitive sequence in the murine and rat protein sequences. The sequence insertions at each site for these two species are not conserved in sequence or length.