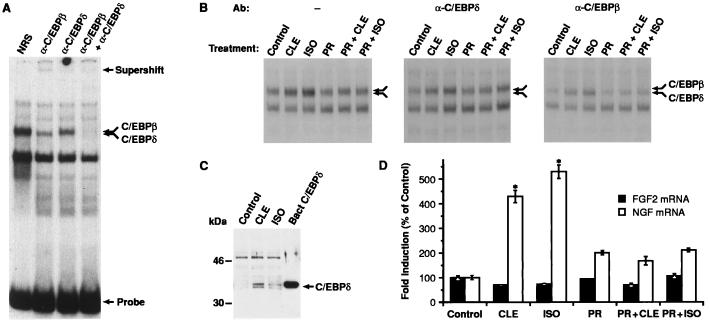
Figure 2.
Analysis of C/EBPδ protein/binding activity in C6–2B glioma cells. (A) Nuclear extracts prepared from C6–2B cells (15) were tested for C/EBP-binding activity by using a 32P-labeled oligonucleotide probe containing the consensus C/EBP site. Samples were preincubated with normal rabbit serum (NRS), α-C/EBPβ, or α-C/EBPδ antisera before addition of probe. The double arrow indicates a doublet corresponding to the C/EBPβ and C/EBPδ complexes. Positions of the supershifted species and free probe are indicated. (B) Cells were exposed for 2 hr to CLE (1 μM) or ISO (1 μM) alone or in combination to PR (1 μM). The double arrow indicates the position of the C/EBPβ-C/EBPδ heterodimers. Preincubation with α-C/EBPδ (Center) or α-C/EBPβ (Right) antisera was carried out to reveal the specific induction of each component of the complex. (C) Example of Western blot analysis of C/EBPδ expression. Ten micrograms of nuclear extracts from CLE- or ISO-treated cells was electrophoresed on a 12% SDS/PAGE gel. Proteins then were transferred to Immobilon P membrane and probed with α-C/EBPδ antibody (1:8,000). Protein/antibody complexes were visualized by using the enhanced chemiluminescence system (ECL, Amersham). Bacterial C/EBPδ was used as positive control. Similar data have been replicated three times. (D) Analysis of NGF mRNA levels in C6–2B cells exposed to CLE or ISO for 3 hr. Data are the mean ± SEM of three separate experiments, with two independent samples per group in each experiment. ∗, P < 0.01 vs. control (ANOVA and Dunnett’s test).