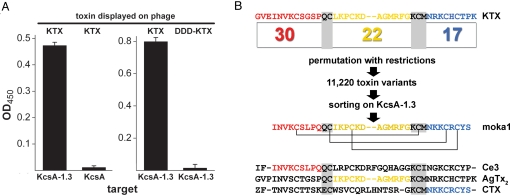
Fig. 1.
KcsA-1.3 channels bind KTX phage and isolate moka1 phage from an α-KTx scaffold library. Phage preparation, library construction, sorting and ELISA protocols, and KcsA, KcsA-1.3, and toxin synthesis and purification are described in SI Materials and Methods. Single-letter codes for amino acids are standard with Z for pyroglutamate. (A) ELISA shows KTX phage bind to KcsA-1.3 but not wild-type KcsA channels (Left) and phage expressing KTX bind to KcsA-1.3 channels, whereas those expressing DDD-KTX do not (Right). Ninety-six well plates coated with KcsA-1.3 or KcsA were incubated with phage (108–1010/well). Data are mean ± SE. for three wells. (B) Library construction and sorting. Thirty-one scorpion toxins that share the α-KTx scaffold were aligned to define three domains (A, B, and C). In KTX, domains were from residues Gly1-Pro12 (A), Leu15-Gly26 (B), and Asn30-Lys38 (C). Domains were linked by sharing of nucleotide codes for amino acids QC (A and B) or KCM (B and C), thereby conserving the important KTX residue Lys27. This yields 30, 22, and 17 unique A, B, and C domains, respectively, and a calculated library diversity of 11,220. Moka1 (GQ153941), a unique toxin isolated from the library, is composed of residues present in the natural toxins Ce3 (red), AgTx2 (yellow), and CTX (blue). Móka is a Hungarian word that translates into English as fun. Isolation of two or more identical clones in 20 by sorting was the basis for further study because the probability of this in the absence of enrichment (e.g., randomly) is smaller than 10−8.