Fig. 1.
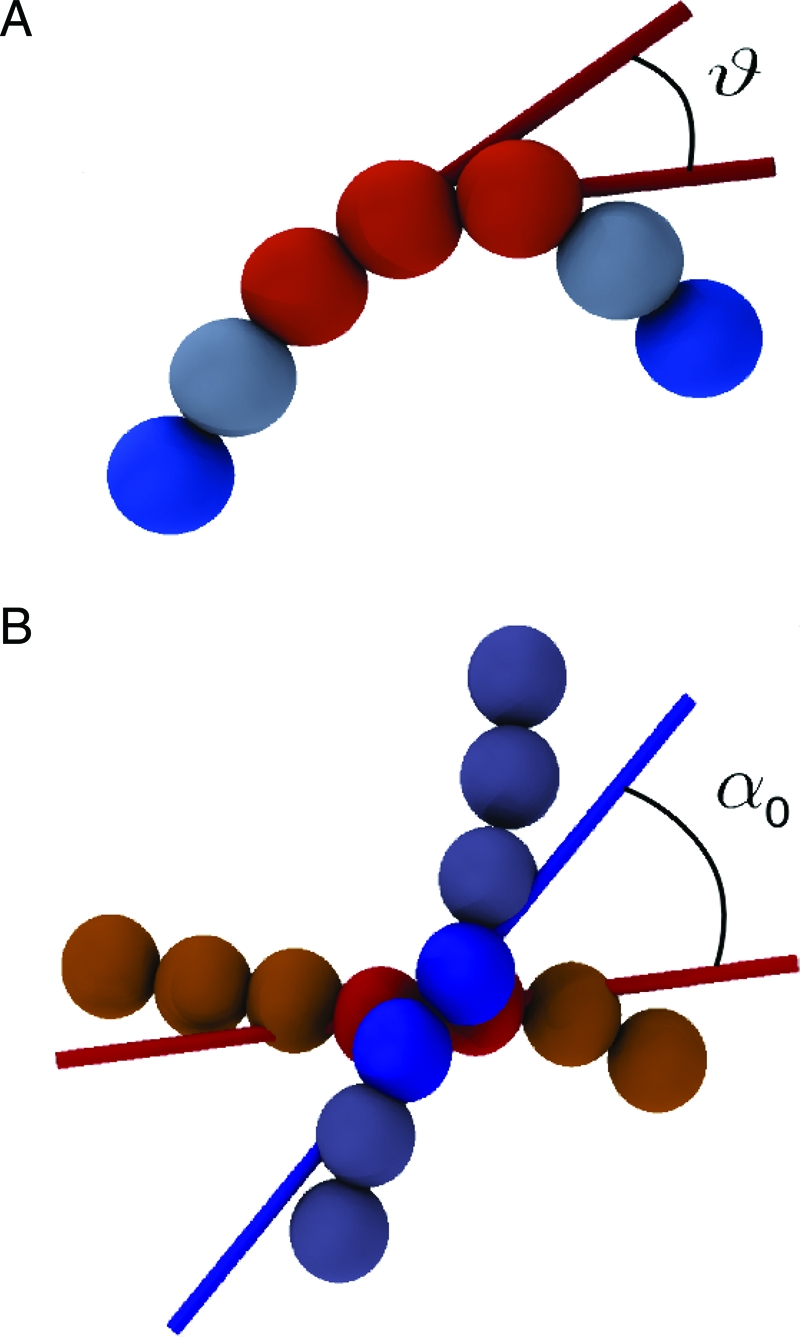
DNA is modeled as a semiflexible chain of beads with diameter equal to 2.5 nm. The flexural rigidity of the DNA molecule is treated by introducing a potential energy term that disfavors bent-chain configurations. As shown in A, the local degree of bending is measured through the angle θ formed by the virtual bonds that join the centers of consecutive beads. As shown in B, a cholesteric interaction between contacting portions of DNA is introduced by favoring a preferential twist angle, α0, between virtual bonds that have a close spatial separation. The figure highlights the pairs of consecutive beads (shown as spheres) bridged by the virtual bonds. The directionality of the latter is shown with colored sticks. The twist angle is calculated accounting for the nonoriented character of dsDNA.
