Abstract
Rhizobial FixK-like proteins play essential roles in activating genes for endosymbiotic life in legume root nodules, such as genes for micro-oxic respiration. In the facultative soybean symbiont, Bradyrhizobium japonicum, the FixK2 protein is the key player in a complex regulatory network. The fixK2 gene itself is activated by the 2-component regulatory system FixLJ in response to a moderate decrease of the oxygen tension, and the FixK2 protein distributes and amplifies this response to the level of approximately 200 target genes. Unlike other members of the cAMP receptor protein family, to which FixK2 belongs, the FixK2 protein does not appear to be modulated by small effector molecules. Here, we show that a critical, single cysteine residue (C183) near the DNA-binding domain of FixK2 confers sensitivity to oxidizing agents and reactive oxygen species. Oxidation-dependent inactivation occurs not only in vitro, as shown with cell-free transcription assays, but also in vivo, as shown by microarray-assisted transcriptome analysis of the FixK2 regulon. The oxidation mechanism may involve a reversible dimerization by intermolecular disulfide-bridge formation and a direct, irreversible oxidation at the cysteine thiol, depending on the oxidizing agent. Mutational exchange of C183 to alanine renders FixK2 resistant to oxidation, yet allows full activity, shown again both in vitro and in vivo. We hypothesize that posttranslational modification by reactive oxygen species is a means to counterbalance the cellular pool of active FixK2, which would otherwise fill unrestrictedly through FixLJ-dependent synthesis.
Keywords: CPR/FNR, gene regulation, nitrogen fixation, nodules, rhizobia
The cAMP receptor protein (CRP)/fumarate-nitrate reductase regulator (FNR) family comprises transcription factors that mainly act as activators in a wide range of bacteria (reviewed in ref. 1). In all cases studied, the active form consists of homodimeric proteins in which each monomer contains an N-terminal sensor domain linked to the C-terminal DNA binding domain via a long α-helix that causes dimerization. These regulators control expression of specific sets of genes implicated in a broad spectrum of processes such as oxidative stress response, micro-oxic and anoxic metabolism, carbon catabolism, and stationary phase survival. The response to the respective environmental or intracellular stimuli is usually transduced through an interaction between a signaling molecule and the sensory domain, whereby a conformational change is induced that leads to the binding of the active dimer to operators near the promoters of the target genes (reviewed in ref. 2). Three modes of signal perception have been described: (i) direct perception of a stressor, as in the Lactobacillus casei FLP protein (3); (ii) dependency on a prosthetic group such as a [4Fe-4S]2+ cluster or heme in Escherichia coli FNR (4) or Rhodospirillum rubrum CooA (5), respectively; and (iii) binding of a small effector molecule like cAMP for E. coli CRP (6) or 2-oxoglutarate for cyanobacterial NtcA (7).
In those CRP/FNR-like proteins that function as oxygen sensors or respond to oxidative stress, the conformational change is either induced via a [4Fe-4S]2+ cluster bound to a cysteine-rich motif or by a dithiol-disulfide switch (reviewed in refs. 1 and 8). There are characteristic differences with respect to the position of the critical cysteine residues. E. coli FNR and similar proteins from Gram-negative bacteria have an iron-sulfur cluster bound to 4 cysteines at the N terminus. In contrast, the iron-sulfur cluster-binding cysteines in FNR-like proteins of Gram positive bacteria (Bacillus subtilis, Bacillus licheniformis) are located at the C terminus (9, 10). Lactococcus lactis FlpA employs not only cysteines but also histidines as ligands for iron-sulfur cluster binding (11). A unique case is that of L. casei FLP (3), in which activation in response to oxygen or oxidative stress occurs by reversible formation of an intramolecular disulfide bond, similar to that of the antioxidant defense protein OxyR (12). Conversely, B. licheniformis ArcR, another CRP/FNR-like protein in that bacterium, is inactivated in vitro via an intermolecular disulfide bridge, but the exact nature of ArcR-mediated control in vivo is not known (13). Finally, E. coli YeiL is a protein for which it is not clear whether it binds a [4Fe-4S]2+ cluster or functions by a dithiol-disulfide switch (14).
Rhizobial FixK-like proteins belong to the CRP/FNR superfamily and play an essential role in activating genes required for the micro-oxic lifestyle either in free-living conditions or in root-nodule symbiosis with leguminous host plants. FixK lacks an obvious oxygen sensory module and is thus integrated in, and cooperates with, more complex regulatory circuits. During the establishment of a symbiosis, rhizobia are not only exposed to a strongly decreased free oxygen concentration but also to transient bursts of reactive oxygen species (ROS) produced by the plants (15). In Bradyrhizobium japonicum, the nitrogen-fixing root-nodule endosymbiont of soybean (Glycine max), a FixK-like protein called FixK2 acts as the key distributor of the low-oxygen signal perceived at the level of the hierarchically superimposed FixLJ 2-component regulatory system. Among the many FixK2 target genes are the fixNOQP and fixGHIS operons for the high-affinity cbb3-type terminal oxidase of micro-oxic bacteroid respiration inside root nodules (16).
Recently, the FixK2 regulon was unraveled by using a transcriptomics approach (17). Also, DNA binding site predictions together with a FixK2-dependent in vitro transcription assay has identified 11 direct target genes or operons for FixK2. The latter studies, carried out with purified FixK2 protein, showed that FixK2 is apparently sufficient to activate transcription in vitro without any identified effector (18). This is puzzling in view of the facts described here that all CRP/FNR-like proteins can be positively or negatively modulated in their activity through bound cofactors or intrinsic, reactive amino acids. Findings reported here now show that posttranslational control occurs at FixK2, whereby a critical cysteine at position 183 in the polypeptide chain is a target for oxidation. This provides a second, important means of affecting FixK2 activity, in addition to the regulation of its expression by FixLJ.
Results
Single Cysteine Mediates Dimerization by Oxidation of FixK2 in Vitro.
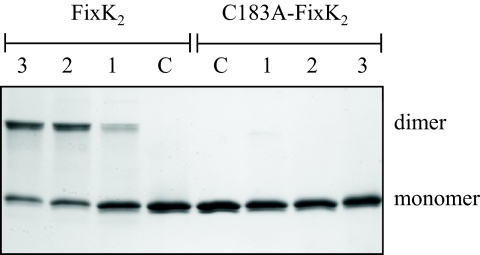
Transcription factors of the CRP/FNR superfamily are usually active as noncovalently assembled dimers. In denaturing SDS/PAGE gels under reducing conditions, FixK2 dimers dissociate and run as monomers (27,785 Da). In nonreducing SDS/PAGE gels, however, we observed stable dimer formation (55,570 Da) after exposure of FixK2 to oxidizing agents. For example, treatment with CuCl2 (0.1–10 mM) and subsequent incubation for 2 h at 25 °C led to the formation of intermolecularly cross-linked dimers whereas nontreated FixK2 migrated as monomers (Fig. 1, 4 left lanes). Similar results were obtained with other oxidizing agents such as ammonium persulfate (APS; 0.05 mM) and H2O2 (0.1 mM), although the efficiencies of dimerization (measured by densitometric analysis of the bands) were only 57% and 41% compared with that after treatment with CuCl2 (10 mM). The FixK2 amino acid sequence contains only one cysteine residue (C183), which might be responsible for the formation of a covalent intermolecular disulfide bond. We therefore constructed a C183A mutant variant of FixK2 and found that it was indeed resistant to oxidation as witnessed by the lack of dimer formation (Fig. 1 Right).
Fig. 1.
Oxidation-induced monomer-dimer switch in FixK2 viewed by SDS/PAGE. The lanes were loaded with 3.5 μM of FixK2 or C183A-FixK2 protein that had been incubated for 2 h at 25 °C with CuCl2 concentrations of 0.1 mM (lanes 1), 1 mM (lanes 2), 10 mM (lanes 3). Nontreated protein samples were run as control (C).
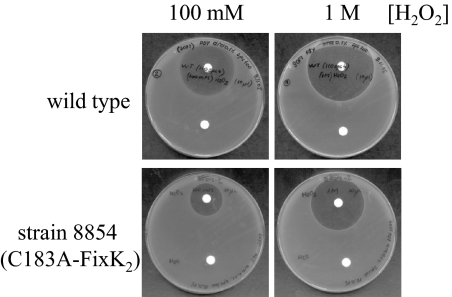
Fig. 4.
H2O2 sensitivity of the B. japonicum WT and the C183A-FixK2-expressing mutant strain 8854. Soft agar (0.9%) plates with PSY medium were inoculated with the indicated B. japonicum strains. Exposure to H2O2 occurred by radial diffusion from the paper disks (placed on top) that had been soaked with the indicated H2O2 concentrations. Disks at the bottom contained H2O for control.
Oxidation of Cysteine 183 Causes Inactivation of FixK2 Function in Vitro.
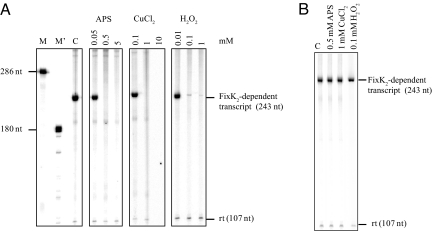
Cysteine 183 of FixK2 is located at the beginning of the predicted helix-turn-helix motif for DNA binding. Mutations of that residue might therefore have been expected to affect transcription activation activity of FixK2; however, this was not the case. When tested in vitro in a transcription activation assay (Fig. 2), the C183A variant of FixK2 proved to be fully active not only in the absence but also in the presence of oxidizing agents (0.5 mM APS, 1 mM CuCl2, 0.1 mM H2O2; Fig. 2B). In contrast, no FixK2-dependent transcripts were synthesized when the WT protein was treated with the same concentrations of the oxidizing agents (Fig. 2A). This suggests that C183 per se is not essential for FixK2 activity but that oxidation at this amino acid renders FixK2 inactive.
Fig. 2.
Transcription activation in vitro by FixK2 and C183A-FixK2 in the presence or absence of oxidizing agents. Template plasmid pRJ8816 containing the FixK2-dependent fixN promoter cloned upstream of a strong transcription terminator was used for multiple-round in vitro transcription with 1.25 μM of purified FixK2 proteins and RNA polymerase holoenzyme from B. japonicum. FixK2 and C183A-FixK2 were incubated with different concentrations of DTT, APS, CuCl2 for 2 h at 25 °C, or with H2O2 for 4 h at 25 °C before the assays; concentrations (in mM) are shown (Top). Assays carried out with FixK2 (A) and C183A-FixK2 (B). Control reactions were performed with nontreated protein (lane C in both panels). Transcripts synthesized in the presence of [α-32P]-UTP were separated on a 6% denaturing polyacrylamide gel and visualized by PhosphorImager analysis of the dried gel. The positions of the FixK2-dependent transcript, the FixK2-independent vector-encoded reference transcript (rt), and 2 RNA size markers (M and M') are marked (nt, nucleotides).
Reversible and Sacrificial Oxidation.
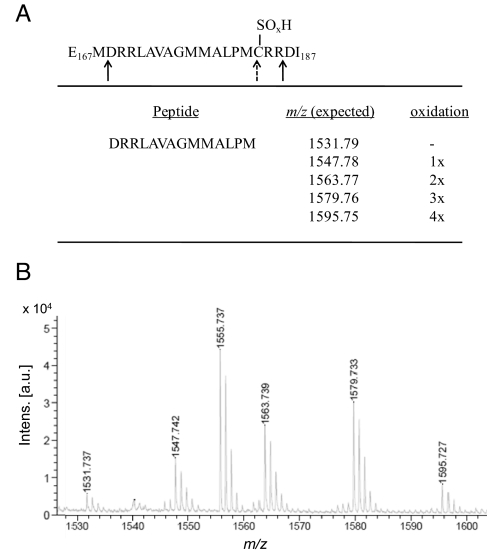
We noticed that FixK2 dimer formation and activity loss caused by CuCl2 were reversible by reduction (i.e., DTT treatment) whereas H2O2-mediated effects were irreversible. We suspected, therefore, that the C183 thiol can not only be oxidized to become a disulfide bridge, but can be further oxidized irreversibly into sulfinic (−SO2H) or sulfonic (−SO3H) acid derivatives. To test this, H2O2-treated FixK2 was run in an SDS/PAGE gel, the band corresponding to the monomer was cut out, and the protein was digested with the peptidyl-Asp endopeptidase from Pseudomonas fragi (AspN). This enzyme not only cleaves peptide bonds N-terminal to aspartic acid but also recognizes cysteine-SO2H and cysteine-SO3H, but not cysteine-SH, as cleavage sites (19). The resulting peptides were analyzed by MS. In fact, AspN digestion of H2O2-treated FixK2 yielded shorter peptides (D169RRLAVAGMMALPM182 of m/z 1531.737, plus oxidized derivatives thereof with 1, 2, 3, or 4 additional oxygen atoms; Fig. 3) instead of the full-length peptide (D169RRLAVAGMMALPMCRR185; Fig. 3A). This suggests that not only the cysteine thiol but also 3 adjacent methionine residues (compare Fig. 3A) are prone to oxidation, probably resulting in methionine sulfoxides or sulfones. Unfortunately, this complication was also the reason why we could not unequivocally identify the precise chemical nature of the type of oxidation that occurred at C183. The AspN-mediated cleavability of H2O2-treated FixK2 at this position allows only the conclusion that the C183 thiol has been converted to either a sulfinic or a sulfonic acid derivative.
Fig. 3.
Strategy to map over-oxidation of cysteine 183 in FixK2. (A) Schematic representation of the AspN endopeptidase digestion strategy. The AspN cleavage sites are indicated with solid arrows. Oxidation of C183 to sulfinic or sulfonic acid introduces an additional cleavage site (dashed arrow, SOxH; x = 2 or 3). (B) MS of peptides after digestion of H2O2-treated FixK2 with AspN. The peptide produced by the extra cleavage at the oxidized cysteine 183 (D169RRLAVAGMMALPM182) was found in different oxidation states (nonoxidized, mono-, di-, tri- or tetra-oxidized). The 1,555.737 peak was identified as peptide D90VFGLESGPSHRLAA104, which resulted from an unspecific cleavage of AspN at the carboxy end.
Analysis of the in Vivo Status of the FixK2 Protein.
The steady-state levels of FixK2 in B. japonicum WT cells, examined by immunoblot analysis, was found to be quite constant, irrespective of the oxic, micro-oxic, or anoxic mode of growth (see Fig. S1, lanes 1–3). This result was unexpected, because previous studies showed a clear induction of fixK2 gene expression in response to decreased oxygen concentrations, although a basal level of fixK2 expression had been noticed even in aerobiosis (16, 17, 20). Implications of this finding will be addressed in the Discussion. Using defined amounts of purified FixK2 as reference, we estimated that one cell with an assumed volume of 1 μm3 contains approximately eight molecules of FixK2, which corresponds to a concentration as low as 10 nM of monomer. By comparison, the estimated cellular concentration of FNR monomer in anoxically grown E. coli was much higher (3.75 μM) (21). FixK2 dimer formation in vivo was never observed, not even after CuCl2 treatment of cells. Because of the minute amount of FixK2 per cell, we did not attempt to show if direct cysteine oxidation occurred in vivo after H2O2 treatment. Therefore, we took the following more indirect measures to demonstrate H2O2 responsiveness in cells.
FixK2 is Involved in H2O2 Stress Response in Vivo.
As FixK2 activity is affected in vitro by H2O2, we tested if B. japonicum resistance or sensitivity to this stressor might involve FixK2. This was done with a qualitative filter disc assay in which growth inhibition by H2O2 of the B. japonicum WT was compared with that of a fixK2 mutant (strain 8854, carrying the C183A variant of FixK2). As shown in Fig. 4, growth inhibition by H2O2 was much more distinct in the WT than in the 8854 strain (respective inhibition zones of 39 mm and 20 mm diameters at 100 mM H2O2, for example). A ΔfixK2 mutant (strain 9043) (16) was also tested and found to be more sensitive than the WT (inhibition zone of 60 mm diameter at 100 mM H2O2). The results may be interpreted to reflect a dual role of FixK2 in oxidative stress sensitivity (WT vs. 8854 comparison) and oxidative stress protection (WT vs. 9043 comparison).
FixK2 Target Gene Expression Changes in Response to H2O2 Treatment.
To further elucidate the oxidative stress sensitivity of FixK2 in vivo, a global microarray analysis of B. japonicum in response to H2O2 was performed. RNA template for cDNA synthesis was isolated from untreated WT cells, and from WT cells treated for 5, 10, and 30 min with 2 mM H2O2, a concentration that did not inhibit growth. The cells had been cultivated under micro-oxic conditions to allow activation of the FixLJ-FixK2 cascade and expression of its associated regulons (20). Changes in the gene expression profiles were compared with the transcription profiles of all known genes in the FixK2 regulon (17), and particularly to the bona fide direct FixK2 targets (17, 18). We found that the majority of genes of the FixK2 regulon decreased in expression after treatment with H2O2. This is visualized in Fig. S2 where the pattern of FixK2 target gene expression in a ΔfixK2 mutant (strain 9043) (16) is displayed next to that of WT cells treated with H2O2 for different time periods. The complete dataset is given in Table S1. A closer examination of the 203 micro-oxically induced genes whose expression exclusively depends on activation by FixK2 revealed a remarkable overlap: after 5, 10, and 30 min of H2O2 treatment, 112, 116, and 186 of these genes were decreased in expression, respectively, corresponding to 55%, 57%, and 92% of the FixK2 targets. Even more convincing was the finding that expression of all those genes previously proven as direct targets for activation by FixK2 (17, 18) were inhibited in response to H2O2 stress (Table 1).
Table 1.
In vivo response of direct FixK2 targets to treatment with 2 mM H2O2
| Gene no. | Gene name | Known or predicted gene product | Mutant 8854 C183A-FixK2 vs. WT | WT treated vs. untreated (H2O2 exposure) |
Mutant 8854 C183A-FixK2 treated vs. untreated (H2O2 exposure) |
||||
|---|---|---|---|---|---|---|---|---|---|
| 5 min | 10 min | 30 min | 5 min | 10 min | 30 min | ||||
| blr2763 | fixN | cbb3-type cytochrome oxidase | — | — | — | −6.25 | — | — | — |
| blr2767 | fixG | Biogenesis of cbb3-type oxidase | — | — | — | −4.85 | — | — | — |
| blr6062 | cycS | Cytochrome c6 | −2.10 | −3.67 | −3.61 | −7.69 | — | — | −2.90 |
| bll7086 | hemN2 | Anaerobic coproporphyrinogen III oxidase | −2.38 | −2.83 | −3.36 | −7.52 | — | — | — |
| bll3998 | Succinate-semialdehyde dehydrogenase | — | −2.32 | −2.02 | −5.56 | — | — | −2.69 | |
| blr4655 | ppsA | Phosphoenolpyruvate synthase | −2.40 | −3.09 | −3.42 | −8.77 | — | — | — |
| bll6073 | phbC | Poly-β-hydroxybutyrate polymerase | — | −2.1 | −2.52 | −7.14 | — | — | — |
| blr6070 | Alcohol dehydrogenase | — | — | — | −4.12 | — | — | — | |
| bll6061 | fixK1 | CRP/FNR-type transcription regulator | −2.49 | −2.23 | −2.44 | −7.30 | — | — | — |
| blr4637 | HspC2 small heat shock protein | — | −3.72 | — | −6.85 | — | — | — | |
| bsr7087 | Hypothetical protein | — | −2.49 | −3.66 | −8.40 | — | −2.98 | −3.53 | |
Data presented in fold-change; changes between +2 and −2 were considered as no change (—). All strains were grown under micro-oxic conditions.
Gene Activation by C183A-FixK2 in Vivo Is More Tolerant to H2O2.
A similar experiment as that described in the preceding paragraph was also carried out with the B. japonicum C183A-FixK2 mutant (strain 8854). Only a few of the 203 FixK2 targets were differentially down-regulated in cells stressed with H2O2 for 5, 10, and 30 min compared with nonstressed cells of strain 8854, i.e., 5 (2%), 17 (8%), and 15 genes (7%). The expression of all other FixK2-dependent genes remained unaffected (Table S1). Likewise, expression of the known, direct FixK2 target genes was less affected by H2O2 treatment in the C183A-FixK2 mutant compared with the WT (Table 1), i.e., transcription of 8 of the 11 targets did not change at all in response to H2O2, and the other 3 targets changed expression only moderately (−3.5-fold at most) and also later (after 10 min) than in the WT. In conclusion, FixK2 does play a role in vivo in the response of cells to oxidative stress.
The following observations substantiated and expanded this conclusion. (i) Expression of most of the direct FixK2 targets were not or only marginally altered in untreated strain 8854 compared with the WT (Table 1, first data column). Likewise, expression of the direct targets is unaltered in oxically grown strain 8854 compared with oxically grown WT. (ii) When the transcriptome of micro-oxically grown strain 8854 was compared with that of the WT grown under the same condition, we noticed several differentially expressed genes that were also differentially expressed in the H2O2-stressed WT compared with untreated WT. Among them were 2 genes with 8.7- and 5.9-fold increased expression, ahpC and ahpD, coding for alkyl hydroperoxide reductase, an H2O2 detoxification enzyme (22).
Discussion
Here, we highlight the peculiar properties of the B. japonicum transcription factor FixK2, and put forward ideas on why the posttranslational modification might benefit free-living and symbiotic cells in the response to oxidative stress.
FixK2, a CRP/FNR Family Member in a Class of its Own.
Members of the CRP/FNR superfamily of transcription factors are predicted to have similar tertiary structures but can be distinguished by their distinct regulatory mechanisms and specificities for different effectors. To activate transcription, CRP/FNR members undergo an allosteric change that is induced either by direct perception (e.g., Lactobacillus casei FLP) (3) or by the binding of a small effector molecule. For example, cAMP induces a conformational change from a dimeric inactive to a dimeric active form in E. coli CRP, whereas it is the binding of CO to heme in R. rubrum CooA that triggers a switch from the “off” to the “on” state (5, 6; reviewed in ref. 23). More complex is the mechanism of Desulfitobacterium hafniense CprK, whose activation requires not only a structural change induced by o-chlorophenol binding but also a redox switch (24). Only the reduced variant binds to DNA. The crystal structure of the ligand-free virulence regulator PrfA of Listeria monocytogenes (25) suggests that it might bind a cofactor for activation, but the nature of the cofactor is unknown. Based on the crystal structure of the inactive apo-form of the NO sensor DNR of Pseudomonas aeruginosa, Giardina and coworkers (26) proposed a novel activation mechanism by which apoDNR undergoes a heme-mediated conformational rearrangement of both the sensing and C-terminal domains to switch to the active conformation upon NO binding.
An exception in the CRP/FNR family of regulators seems to be the regulatory protein SdrP of Thermus thermophilus (27), whose crystal structure and in vitro transcription properties suggest that it does not require an effector to bind DNA. B. japonicum FixK2 also does not appear to require a cofactor or a modification to become active (18). An alignment of FixK2 with other CRP/FNR amino acid sequences from Gram-positive and Gram-negative bacteria revealed that, although most of the CRP/FNR-type secondary protein structure elements are retained (Fig. S3), the sensory domain is strikingly different in that only one cysteine residue is present in the entire FixK2 sequence (C183; Fig. S3). Located in the first position of α helix E of the predicted DNA binding domain, C183 is neither conserved in the other 15 CRP/FNR-type transcription factors of B. japonicum (28), nor in any other known CRP/FNR protein. The peculiar position of C183 might be instrumental to efficiently shut down FixK2 activity under any environmental condition that promotes FixK2 oxidation.
Two Modes of FixK2 Oxidation.
Most of the transcription factors involved in the oxidative stress response use reactive cysteines to control their activity, often forming a reversible disulfide bond (29, 30). In contrast, although, the peroxide-sensing regulator OhrR of B. subtilis undergoes oxidation either to a reversible cysteine sulfenic acid intermediate or to cysteine sulfinic and sulfonic acid derivatives (31), modifications that are irreversible (i.e., sacrificial) in bacteria. Our in vitro analyses showed that FixK2 might select between the reversible and irreversible modes of inactivation, depending on the nature of the oxidant. H2O2 primarily led to over-oxidation, whereas CuCl2 oxidized FixK2 to the dimeric, reversible form. One can only speculate that the choice between the 2 modes of control is perhaps crucial under different kinds of stress conditions—copper stress, oxidative stress—that will affect cell survival. Also, one has to consider the cost a cell has to pay in case it needs to degrade irreversibly oxidized FixK2 and resynthesize it thereafter. Because we had failed to demonstrate the formation of disulfide-bridged dimers in vivo, we assume that direct, sacrificial oxidation of C183 to acid derivatives is more likely; yet, the chemical mode of oxidation in vivo remains an unsolved issue.
Possible Relevance of FixK2 Inactivation in Vivo.
Before this work, regulation of fixK gene transcription was the only known means to control formation of FixK-like proteins in cells. In B. japonicum, transcription of fixK2 is activated by FixJ and negatively autoregulated by FixK2 or a FixK2-dependent function (16). In other bacteria, such as S. meliloti and Caulobacter crescentus, a FixT-like protein negatively regulates FixK synthesis either by blocking the FixL autokinase activity (32) or by mimicking and outcompeting FixJ (33). A gene product with functional homology to FixT has not been identified in B. japonicum, and the exact mechanism how FixK2 represses its own synthesis is not known.
Although the transcription of fixK2 is induced in micro-oxically grown cells compared with oxically grown cells (16), immunoblot analysis now showed that the levels of FixK2 protein in vivo do not vary much in cells grown in different conditions (oxic, micro-oxic, anoxic; Fig. S1). While the reason for this surprising discrepancy is not known, the presence of significant amounts of FixK2 even in aerobically grown cells, in which FixK2 target genes are poorly activated, can now be taken as an argument in support of a posttranslational control. The purpose of FixK2 oxidation would then be to prevent an unnecessary activation of the FixK2 regulon in oxic conditions.
Our proposal that FixK2 oxidation indeed occurs in vivo relies on more indirect evidence. For example, most of the FixK2 targets, particularly all of the direct target genes, are inhibited in their expression in response to oxidative stress. Their expression decreased even more strongly in a ΔfixK2 strain, with fold-change factors between −7.3 and −111 compared with the WT (Table S1). Such an inhibitory effect was hardly, or not at all, detectable in the C183A-FixK2 mutant, corroborating the importance of that single cysteine. As already argued, inactivation of FixK2 via oxidation of C183 could prevent transcription of FixK2 targets in situations in which such genes are not needed or are even detrimental if expressed. The following other scenarios may be envisaged in this context. (i) During infection, rhizobia encounter transient bursts of ROS (primarily H2O2 and superoxide) that are produced by enzymes in the apoplast as part of the plant defense system (15, 34), and rhizobia must have special means to overcome this threat. However, H2O2 is not merely harmful for infecting bacteria but can be essential for optimal root-nodule development, as shown for the Medicago sativa–Sinorhizobium meliloti symbiosis (35, 36). With ROS signals from the plant, the posttranslational inactivation of FixK2 could prevent a wasteful and energy-consuming transcription of genes not yet needed at early stage of nodulation. To demonstrate such effects experimentally will be difficult, even more so as we did not notice a phenotypic difference between the WT and the C183A-FixK2 mutant in terms of nodule formation and nitrogen fixation activity. (ii) Although mature nodules are thought to be relatively well protected from ROS by enzymes from both symbiotic partners (37), senescent nodules enhance ROS production and concurrently decrease the antioxidant defense (38, 39), leading to oxidative damage of lipids, proteins, and DNA (40). Again, ROS-dependent shut-down of FixK2 activity would assist in down-regulating symbiotic functions that become futile during senescence. (iii) FixK2 controls the fixNOQP-encoded high-affinity cbb3-type oxidase (16) that supports bacteroid respiration in micro-oxic conditions that are prevalent in nodules. As respiration turnover is high, ROS is assumed to be generated as a side product. Likewise, an electron donor for respiration, the flavoenzyme NADH dehydrogenase II, produces ROS (41). In case of excessive electron flow, the generation of ROS will inactivate FixK2 and subsequently stop the production of the cbb3 oxidase and, hence, the generation of more ROS. This kind of negative autoregulation guarantees a good balance between the beneficial and detrimental effects of bacteroid respiration. Regulating FixK2 posttranslationally might have at least 2 advantages: (i) it could be a means to control its activity rapidly and (ii) a pool of active FixK2 might be regenerated quickly by reduction, unless its critical cysteine has been over-oxidized.
The role of a CRP/FNR-type protein in the adaptation to H2O2 is not unprecedented. Zeller and coworkers (42) reported for Rhodobacter sphaeroides that FnrL functions not only as a key regulator but also initiates the shift toward high-oxygen metabolism. Unlike FixK2, however, FnrL contains a [4Fe-4S]2+ cluster that is sensitive to destruction by O2 and ROS, and so the expression of many genes whose expression depends on FnrL is decreased. When the [4Fe-4S]2+ cluster of FnrL is reestablished on FnrL, expression of the FnrL-dependent genes is restored. In addition, transcription of fnrL is induced upon H2O2 treatment.
In contrast to the observation with fnrL, we noticed that the expression of fixK2 itself decreases during treatment with H2O2 (see gene bll2757 in Table S1). This supports the idea of a sequential regulation, involving an initial, fast response that leads to inactivation of FixK2 by oxidation at C183, after which the expression of fixK2 is slowly decreased, possibly by cessation of the FixLJ-mediated activation. The exposed heme of FixL seems to be a likely target for attack by ROS.
Strain 8854 carrying the C183A-FixK2 variant is more resistant to H2O2 than the WT, also in contrast to the ΔfixK2 mutant, which exhibits higher sensitivity. This peculiar result raises the question whether FixK2 is partly involved in the control of ROS-protective genes. Under micro-oxic growth conditions, the products of such protection genes might quench small amounts of ROS. As ROS increases to higher levels, FixK2 itself would be quickly inactivated to prevent further accumulation of ROS from the terminal oxidase, for instance. The observed pattern of moderate stress gene expression in strain 8854 led us to assume that this strain is prestressed and therefore has a certain advantage at an emerging oxidative stress over a strain that first needs to induce the expression of protection genes. The observed higher level of ahpCD gene expression, for example, could account for the higher tolerance toward H2O2 in strain 8854.
In summary, our data appear to have helped to fill a gap in understanding how the FixLJ-FixK2 cascade works. As this cascade was hitherto thought to comprise only the two sequentially acting activators (FixJ, FixK2), the unlimited activation of their target genes had to be regarded as an unlikely scenario. What was previously missing was at least one negatively acting check-point that transmits a negative feedback to the level of the FixK2 protein, depending on the cellular status. The oxidative stress response of FixK2 is shown to play precisely this role.
Materials and Methods
Bacterial Strains and Growth Conditions.
E. coli strains used in this work were DH5α and BL21 (DE3). B. japonicum strains used in this work were 110spc4 (WT; 43), 9043 (ΔfixK2; 16), and 8854 (C183A-FixK2 derivative). Details of growth conditions can be found in the SI Text.
Plasmid and Strain Constructions.
For plasmids and strain constructions, see the SI Text.
Biochemical Methods.
Full details and associated references of the following biochemical methods can be found in the SI Text: Protein expression and purification, biochemical characterization of FixK2 protein derivatives, in-gel digestion, MS analyses, and immunoblotting.
In Vitro Transcription Experiments.
Multiple-round in vitro transcription assays were performed as described in ref. 18 with the exception that purified FixK2 derivatives (WT and mutant) were treated as indicated before testing them in the assays. For details, see the SI Text.
H2O2 Sensitivity.
For zone inhibition assays, cells were grown aerobically, washed twice with peptone-salts-yeast extract (PSY) medium, and used to inoculate 15 mL prewarmed (42 °C) PSY soft agar (0.9% agar) to an OD600 of 0.04. In parallel, samples were taken to determine colony-forming units by plating serial dilutions. Sterile filter disks containing 10 μL of H2O2 (10 mM to 1 M) were placed on the plate. As control, 10 μL H2O was used. The plates were incubated at 30 °C, and the diameter of the growth inhibition zone was determined after 3 to 4 days of incubation.
RNA Isolation, cDNA Synthesis, and Hybridization to Microarrays.
WT and strain 8854 were grown micro-oxically in 25-mL cultures until midexponential phase. Cells were then exposed to 2 mM H2O2 for different time periods. Harvest and storage of cells and subsequent RNA extraction were done as described in ref. 44. RNA was then purified, reverse-transcribed to cDNA, processed to fragments, and hybridized to a whole-genome B. japonicum Affymetrix GeneChip (44). For each strain, a minimum of three biological replicates was prepared.
Data Processing.
Signal intensity detection, normalization and statistical analysis were done as already established (17, 45). Only those probe sets that were called “present” or “marginal” in ≥67% of the replicates of each experiment were considered for further analysis. The hierarchical clustering of genes was performed as described in ref. 45.
Biocomputing Analysis of FixK2.
For biocomputing analysis of FixK2, see the SI Text.
Supplementary Material
Acknowledgments.
We thank Olivera Volarevic (who, sadly, died in August 2009) for expert technical assistance. The help of Andrea Patrignani, Hubert Rehrauer, Stefan Zoller, Peter Hunziker, and Yolanda Auchli (Functional Genomics Center Zurich) in the microarray and MS analyses is greatly appreciated. Mariette Bonnet (our laboratory) generously provided some of the purified recombinant FixK2 protein samples. This work was supported by grants from the Swiss National Foundation for Scientific Research and ETH Zürich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908097106/DCSupplemental.
References
- 1.Körner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 2.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 3.Gostick DO, Green J, Irvine AS, Gasson MJ, Guest JR. A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology. 1998;144:705–717. doi: 10.1099/00221287-144-3-705. [DOI] [PubMed] [Google Scholar]
- 4.Kiley PJ, Beinert H. The role of Fe–S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 5.Lanzilotta WN, et al. Structure of the CO sensing transcription activator CooA. Nat Struct Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 6.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc Natl Acad Sci USA. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valladares A, Flores E, Herrero A. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2008;190:6126–6133. doi: 10.1128/JB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green J, Crack JC, Thomson AJ, LeBrun NE. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12:145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Cruz Ramos H, et al. Anaerobic transcription activation in Bacillus subtilis: Identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klinger A, Schirawski J, Glaser P, Unden G. The fnr gene of Bacillus licheniformis and the cysteine ligands of the C-terminal FeS cluster. J Bacteriol. 1998;180:3483–3485. doi: 10.1128/jb.180.13.3483-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott C, Guest JR, Green J. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol Microbiol. 2000;35:1383–1393. doi: 10.1046/j.1365-2958.2000.01799.x. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 13.Wohlkönig A, Stalon V, Vander Wauven C. Purification of ArcR, an oxidation-sensitive regulatory protein from Bacillus licheniformis. Protein Expr Purif. 2004;37:32–38. doi: 10.1016/j.pep.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Anjum MF, Green J, Guest JR. YeiL, the third member of the CRP-FNR family in Escherichia coli. Microbiology. 2000;146:3157–3170. doi: 10.1099/00221287-146-12-3157. [DOI] [PubMed] [Google Scholar]
- 15.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 16.Nellen-Anthamatten D, et al. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol. 1998;180:5251–5255. doi: 10.1128/jb.180.19.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesa S, et al. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol. 2008;190:6568–6579. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesa S, Ucurum Z, Hennecke H, Fischer HM. Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2. J Bacteriol. 2005;187:3329–3338. doi: 10.1128/JB.187.10.3329-3338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner E, et al. A method for detection of overoxidation of cysteines: Peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sciotti MA, Chanfon A, Hennecke H, Fischer HM. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum. J Bacteriol. 2003;185:5639–5642. doi: 10.1128/JB.185.18.5639-5642.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutton VR, Mettert EL, Beinert H, Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panek HR, O'Brian MR. KatG is the primary detoxifier of hydrogen peroxide produced by aerobic metabolism in Bradyrhizobium japonicum. J Bacteriol. 2004;186:7874–7880. doi: 10.1128/JB.186.23.7874-7880.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan MK. CooA, CAP and allostery. Nat Struct Biol. 2000;7:822–824. doi: 10.1038/79559. [DOI] [PubMed] [Google Scholar]
- 24.Levy C, et al. Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol Microbiol. 2008;70:151–167. doi: 10.1111/j.1365-2958.2008.06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eiting M, Hageluken G, Schubert WD, Heinz DW. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol Microbiol. 2005;56:433–446. doi: 10.1111/j.1365-2958.2005.04561.x. [DOI] [PubMed] [Google Scholar]
- 26.Giardina G, Rinaldo S, Castiglione N, Caruso M, Cutruzzolà F. A dramatic conformational rearrangement is necessary for the activation of DNR from Pseudomonas aeruginosa Crystal structure of wild-type DNR. Proteins. 2009;77:174–180. doi: 10.1002/prot.22428. [DOI] [PubMed] [Google Scholar]
- 27.Agari Y, Kashihara A, Yokoyama S, Kuramitsu S, Shinkai A. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol Microbiol. 2008;70:60–75. doi: 10.1111/j.1365-2958.2008.06388.x. [DOI] [PubMed] [Google Scholar]
- 28.Mesa S, Hennecke H, Fischer HM. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem Soc Trans. 2006;34:156–159. doi: 10.1042/BST0340156. [DOI] [PubMed] [Google Scholar]
- 29.Kiley PJ, Storz G. Exploiting thiol modifications. PLoS Biol. 2004;2:e400. doi: 10.1371/journal.pbio.0020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Soonsanga S, Lee JW, Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol Microbiol. 2008;68:978–986. doi: 10.1111/j.1365-2958.2008.06200.x. [DOI] [PubMed] [Google Scholar]
- 32.Garnerone AM, Cabanes D, Foussard M, Boistard P, Batut J. Inhibition of the FixL sensor kinase by the FixT protein in Sinorhizobium meliloti. J Biol Chem. 1999;274:32500–32506. doi: 10.1074/jbc.274.45.32500. [DOI] [PubMed] [Google Scholar]
- 33.Crosson S, McGrath PT, Stephens C, McAdams HH, Shapiro L. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc Natl Acad Sci USA. 2005;102:8018–8123. doi: 10.1073/pnas.0503022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos R, Herouart D, Sigaud S, Touati D, Puppo A. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol Plant-Microbe Interact. 2001;14:86–89. doi: 10.1094/MPMI.2001.14.1.86. [DOI] [PubMed] [Google Scholar]
- 35.Pauly N, et al. Reactive oxygen and nitrogen species and glutathione: Key players in the legume-Rhizobium symbiosis. J Exp Bot. 2006;57:1769–1776. doi: 10.1093/jxb/erj184. [DOI] [PubMed] [Google Scholar]
- 36.Jamet A, Mandon K, Puppo A, Herouart D. H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J Bacteriol. 2007;189:8741–8745. doi: 10.1128/JB.01130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matamoros MA, et al. Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol. 2003;133:499–509. doi: 10.1104/pp.103.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alesandrini F, Mathis R, Van de Sype G, Herouart D, Puppo A. Possible roles for a cysteine protease and hydrogen peroxide in soybean nodule development and senescence. New Phytologist. 2003;158:131–138. [Google Scholar]
- 39.Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 1999;121:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans PJ, et al. Oxidative stress occurs during soybean nodule senescence. Planta. 1999;208:73–79. [Google Scholar]
- 41.Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 42.Zeller T, Moskvin OV, Li K, Klug G, Gomelsky M. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J Bacteriol. 2005;187:7232–7242. doi: 10.1128/JB.187.21.7232-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 44.Hauser F, et al. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol Genet Genomics. 2007;278:255–271. doi: 10.1007/s00438-007-0246-9. [DOI] [PubMed] [Google Scholar]
- 45.Pessi G, et al. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant-Microbe Interact. 2007;20:1353–1363. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.