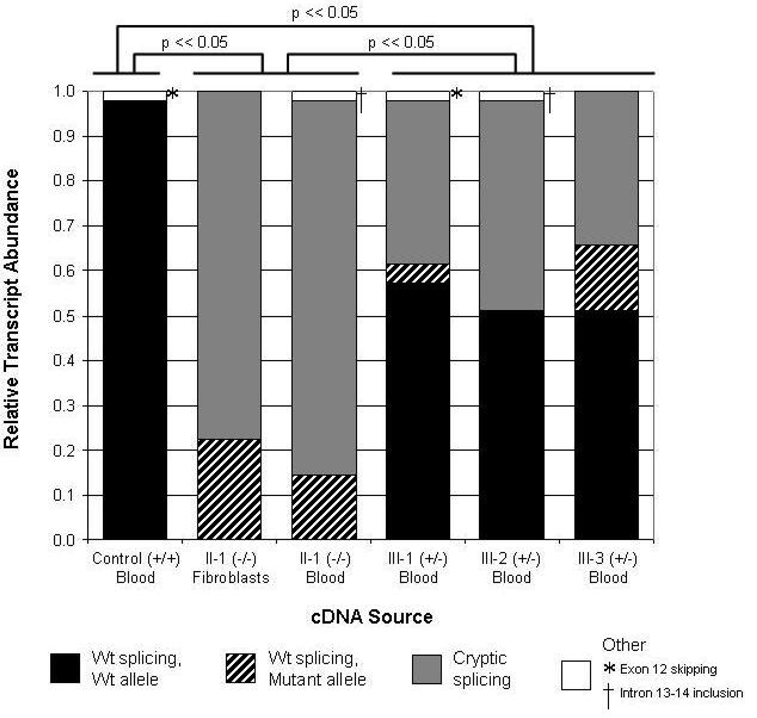
Figure 4.
Quantification of wild-type and mutant cryptic splicing of the PKP2 transcript in six clinical samples. Relative transcript abundance in two sources of RNA from the proband is compared to transcript abundance in RNA derived from an unaffected control and from the proband’s three heterozygous offspring. Proportion of wild-type splicing derived from the wild-type allele (c.2484C) is shaded in black. Proportion of wild-type splicing derived from the mutant allele (c.2484T) is shaded with black diagonal bars. Proportion of cryptic splicing with the 7-bp deletion at the end of exon 12 is shaded in gray. Minor splice forms (see Fig. 5), shaded in white, include exon 12 skipping (*) and intron 13–14 retention (†). Statistical differences in the relative abundance of wild-type and mutant splice forms are indicated above the graph, comparing control, combined proband samples, and combined heterozygous samples.