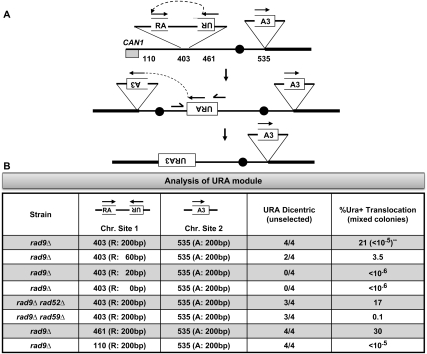
Figure 5.
Synthetic inverted repeats fuse to form dicentrics, and then recombine to form translocations. (A) Three segments of the URA3 gene were cloned into two modules (see Supplemental Fig. S6 for details of module construction and chromosome insertion). The two modules were then inserted into the specific sites in ChrVII as indicated. Inverted repeat recombination joins UR to RA modules via recombination of shared “R” sequences to form a dicentric. A second recombination event joins “A” to “A3” to form an intact URA3 gene, rendering the cells Ura+. (B) Each rad9 strain contains a UR or RA module in one of three sites on the left arm of ChrVII, and all strains contain the A3 module at one site on the right arm. The UR modules differ in the amount of sequence homology shared with the RA module (size of “R” is 200, 60, 20, or 0 bp of homology, as indicated). Each modified rad9 strain was analyzed for the presence of the specific PCR fragment diagnostic of a dicentric chromosome, in four independent cultures, using dicentric URA primers (Supplemental Fig. S9). CanR mixed colonies were generated from each rad9 strain, and the frequency of Ura+ cells in cells from mixed colonies was determined (see the Materials and Methods). (**) The frequency of Ura+ colonies in Cans cells is shown in Supplemental Figure S8.