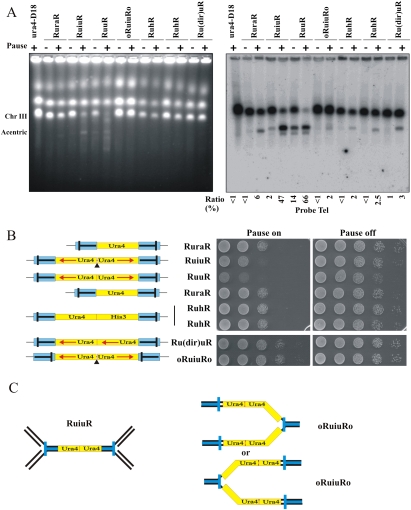
Figure 3.
Fork arrest at inverted repeats also results in large palindromic chromosomes. (A) PFGE of DNA from the indicated strains either without (off; −) or 48 h after (on; +) the induction of rtf1-dependent fork arrest. (Left) Ethidium stained. (Right) Southern blotted with a telomere-proximal probe. The percentage of signal corresponding to the acentric chromosome (1.4 Mb) was calculated (numerals) as a percentage of total signal (2 × 3.5 Mb + 1.4 Mb). (B, left) Schematic. (Blue) RTS1. (Bar in RTS1) The direction that, when a fork approaches, it will be arrested. Red arrows below ura4 (yellow) and his3 (green) indicate direction of transcription. (Solid triangle) 14-bp “interruption”. (Right) Viability analysis in the absence (Pause on) or presence (Pause off) of thiamine to regulate rtf1 for the strains indicated. (C) Schematic of the expected pattern of replication arrest for RuiuR and oRuiuRo. The blue bar indicates the direction that, when a fork approaches, it will be arrested.