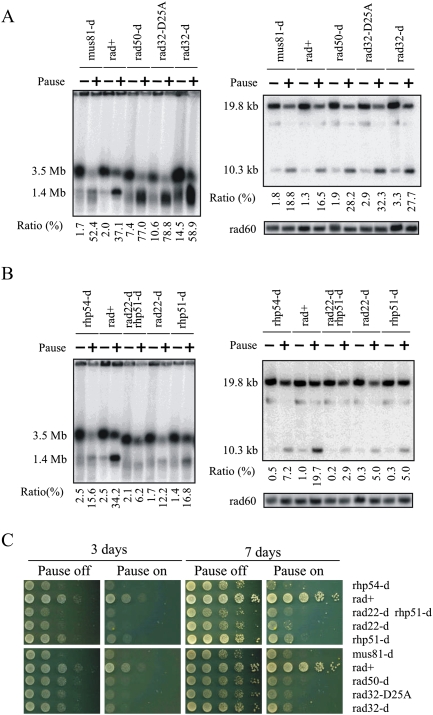
Figure 4.
HR proteins are required for viability and GCR. (A,B) DNA prepared from RuiuR strains indicated, grown either without (off; −) or three generations after (on; +) the induction of rtf1-dependent fork arrest, was analyzed by either PFGE (left) or SalI digestion (right) (see Supplemental Fig. S3C for SalI fragments) and Southern blotting with a telomere-proximal probe. The panel labeled “rad60” represents the same gel probed with a ChrII-specific probe. (Bottom) The ratio of signal corresponding to the acentric chromosome (1.4 Mb or 10.3 kb, respectively, expressed as a percentage) was calculated compared with total signal (2 × 3.5 Mb + 1.4 Mb or 2 × 19.8 kb + 10.3 kb). The rad22Δ rhp51Δ strain was used because rad22rad52 mutants can accumulate suppressors that are Rhp51Rad51-dependent. rad32 = mre11. (C) Viability analysis in the presence (Pause off) or absence (Pause on) of thiamine to regulate rtf1.