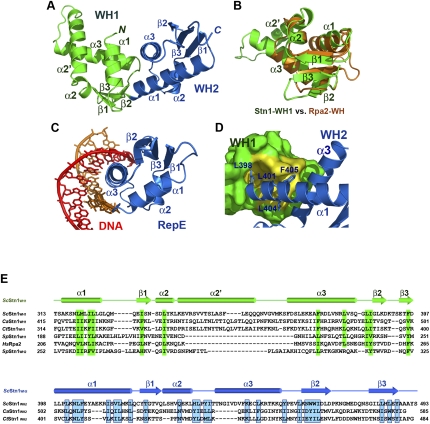
Figure 7.
Crystal structure of the C-terminal domain of S. cerevisiae Stn1. (A) Ribbon diagram of ScStn1C. The WH1 and WH2 motifs of ScStn1N are colored as in Figure 3A. The secondary structure elements are labeled. The dotted line represents the disordered loop (472–479) between strands β2 and β3 of WH2. Although sharing the same topology, the two WH motifs are quite different in structure. (B) Superposition of the WH1 motif of ScStn1 (in green) on the NMR (nuclear magnetic resonance) structure of the WH motif of Rpa2 (in orange) (Mer et al. 2000). Except for a large insertion between α2 and α3, the rest of Stn1WH1 closely resembles the WH motif of Rpa2. (C) Ribbon diagram of the RepE–DNA complex (Komori et al. 1999). The orientation of the WH motif of RepE is the same as the WH2 of ScStn1C in A. (D) The hydrophobic interactions between the WH1 and WH2 motifs of ScStn1C. There are no linker residues between WH1 and WH2, so that ScStnC folds into a globular and compact structure. WH1 is shown in surface representation and is colored in green, except for the WH2-interacting surface, shown in yellow. WH2 is in ribbon representation. Side chains of residues in WH2 important for the WH1–WH2 interaction are shown in stick representations. (E) Amino acid sequence alignment of the C-terminal WH1 and WH2 motifs of budding yeast Stn1 family members together with the WH motifs of S. pombe Stn1 and human Rpa2. The alignment with Rpa2 is based on the NMR structure of the Rpa2C–UNG2 complex (Mer et al. 2000). Secondary structure assignments from our ScStn1C crystal structure are shown. Conserved hydrophobic residues in WH1 and WH2 are highlighted in green and blue blocks, respectively. In contrast to the WH motifs in budding yeasts, both WH1 and WH2 of SpStn1 are similar to Rpa2WH.