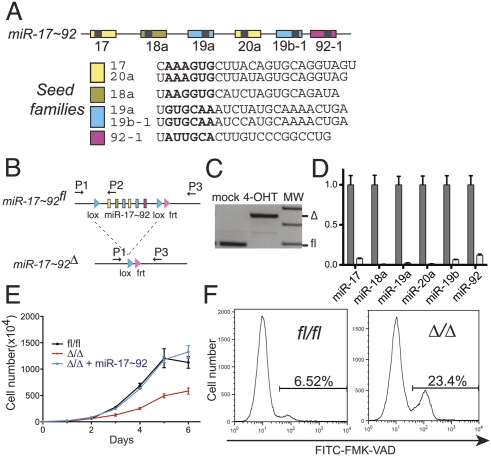
Figure 1.
miR-17∼92 suppresses cell death in Eμ-Myc lymphomas. (A) Schematic representation of the miR-17∼92 cluster. Each miRNA is represented by a colored box and is color-coded based on the seed family to which it belongs. The sequence of each mature miRNA is also shown. (B) Schematic of the conditional miR-17∼92 knockout allele. Arrows represent the primers used to detect the floxed and the deleted (Δ) alleles. (C) PCR on genomic DNA extracted from Eμ-Myc;miR-17∼92fl/fl;Cre-ER lymphoma cells mock-treated or after 4 d of 4-OHT treatment. (D) Quantitative RT–PCR analysis of the expression of miR-17∼92 in lymphoma cells before (gray bars) and after (white bars) 4-OHT treatment. Each component of miR-17∼92 was detected independently, and the results were normalized relative to the expression observed in mock-treated cells. Each experiment was performed in quadruplicate. Error bar, standard deviation (SD). (E) Growth curves of miR-17∼92fl/fl cells (black line), miR-17∼92Δ/Δ cells (red line), and miR-17∼92Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (blue line). Error bars, SD of three replicates. The plot is representative of three independent experiments. (F) Caspase activity in exponentially growing miR-17∼92fl/fl and miR-17∼92Δ/Δ lymphoma cells as detected by flow cytometry using FITC-conjugated VAD-FMK. The percent of VAD-FMK+ cells is shown.