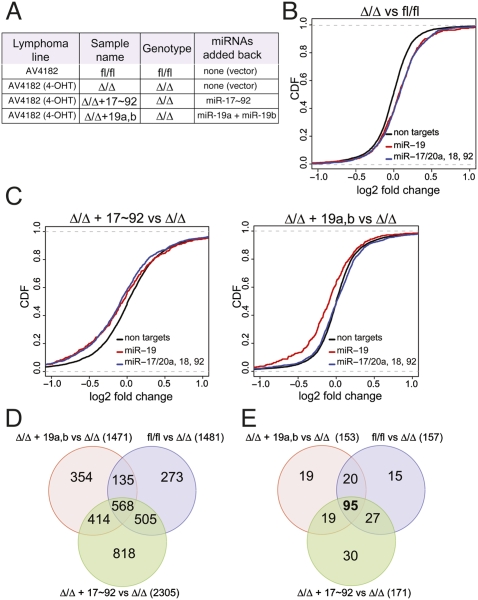
Figure 3.
Gene expression profiling identifies miR-19 targets in Eμ-Myc lymphoma cells. (A) Description of the various lymphoma cells used. (B) Differences in mRNA levels between miR-17∼92fl/fl and miR-17∼92Δ/Δ lymphoma cells transduced with the empty PIG vector were monitored with microarrays. Cumulative distribution function (CDF) plots are shown for mRNAs that do not contain miR-17∼92 seed matches in their 3′UTRs (black line), mRNAs containing one or more seed matches for miR-19 in their 3′UTR (red line), and mRNAs containing one or more seed matches for either miR-17, miR-20a, miR-18a, or miR-92 (blue line). In the absence of endogenous miR-17∼92 expression, a statistically significant up-regulation (P-value < 2.22e-16, KS test) is observed for the predicted miR-17∼92 targets relative to the background gene population. (C) CDF plots of the changes in mRNA expression levels between miR-17∼92Δ/Δ + PIG-miR-17∼92 and miR-17∼92Δ/Δ lymphoma cells (left panel) and between miR-17∼92Δ/Δ + PIG-miR-19a,b and miR-17∼92Δ/Δ lymphoma cells (right panel). (D) Venn diagram summarizing the overlap in gene expression changes observed between the various transduction experiments. (E) As in D, but the analysis was restricted to mRNAs whose 3′UTR contains at least one predicted binding site for miR-19.