Abstract
This article describes a method for the global profiling of the substrate specificities of DNA ligases and illustrates examples using the Taq and T4 DNA ligases. The method combines oligonucleotide arrays, which offer the benefits of high throughput and multiplexed assays, with mass spectrometry to permit label-free assays of ligase activity. Arrays were prepared by immobilizing ternary biotin-tagged DNA substrates to a self-assembled monolayer presenting a layer of streptavidin protein. The array represented complexes having all possible matched and mismatched base pairs at the 3′ side of the nick site and also included a number of deletions and insertions at this site. The arrays were treated with ligases and adenosine triphosphate or analogs of the nucleotide triphosphate and then analyzed by matrix-assisted laser desorption-ionization mass spectrometry to determine the yields for both adenylation of the 5′-probe strand and joining of the two probe strands. The resulting activity profiles reveal the basis for specificity of the ligases and also point to strategies that use ATP analogs to improve specificity. This work introduces a method that can be applied to profile a broad range of enzymes that operate on nucleic acid substrates.
INTRODUCTION
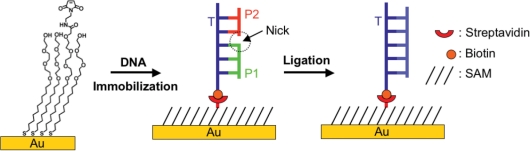
The DNA ligases are a family of enzymes that repair nicked sites within double helical DNA. These enzymes play a fundamental role in the maintenance of organism genomes and have enabled applications in biotechnology (1,2). Individual members of the family vary in the specificities with which they discriminate among substrates and ligases having both high and low discrimination for their substrates have been important in applications (3,4). In the current paper, we report an efficient method for profiling the specificities of ligases. Our approach uses mass spectrometry to characterize the activities of the ligase towards an array of DNA substrates immobilized to a self-assembled monolayer (Figure 1). Mass spectrometry offers the benefit that it can compare yields for each of the multiple steps in the ligase activity and therefore reveal the mechanistic basis for specificity. We illustrate this approach by profiling the specificities of the T4 and Taq DNA ligases and we characterize the enhanced specificity that can be achieved with the use of ATP analogs.
Figure 1.
Scheme showing the strategy used to assemble and assay ternary oligonucleotide substrates on a monolayer.
DNA ligases join two adjacent DNA strands aligned head-to-tail on a DNA template to give a phosphodiester bond between 5′ phosphate and 3′ hydroxyl termini of the strands. Early biochemical studies have revealed that the mechanism of DNA ligation follows three steps (2,5). In the first, the ligase reacts with either NAD+ or ATP to covalently modify an active site lysine residue with adenosine monophosphate (AMP). In the second step, the ligase transfers the AMP moiety to the 5′ phosphate group of the substrate DNA strand. In the third step, the 3′ hydroxyl group of the other strand reacts with the activated strand to give a native phosphodiester linkage with concomitant release of AMP. This mechanism is shared by both NAD+- and ATP-dependent ligases across all biological species (6–12).
Yet, individual ligases show wide differences in their specificities for joining strands that have base pair mismatches at the nick site (2,13–15). Certain ligases are known to exhibit little specificity, including the African swine fever virus (ASFV) DNA ligase, which is among the lowest-fidelity ligases and tolerates mismatches at the 3′ (as well as the 5′) side of the nick site (3). For example, this ligase repairs a nick having a C:T mismatched base pair more efficiently than the correctly matched C:G base pair. In contrast, NAD+-dependent ligases from thermophilic bacteria show a 100-fold better discrimination of matched over mismatched substrates than does the T4 DNA ligase (4).
The methods now used to assay ligase activity—which rely on gel electrophoresis of labeled oligonucleotides—have a limited throughput and are tedious when applied for profiling the activity of the ligase for a family of substrates (16). Efforts to increase throughput have adopted homogeneous phase assays that use a secondary reaction to detect the product of the ligase reaction (17), but the use of labels and multiple steps increase the incidence of false positive and negative results.
Mass spectrometry-based approaches offer the benefits of not requiring labels and avoiding electrophoretic separation of analytes. The former is particularly significant in those cases where the label can interfere with the biochemical activity being assayed and also requires additional steps in the assay. These considerations have motivated the use of mass spectrometry for monitoring enzymatic-modification of DNA in applications including genotyping (18–20), epigenotyping (21,22), and diagnostics (23). When combined with tailored surface chemistries, mass spectrometry can be applied in a high throughput format and is compatible with immobilized arrays. For example, we have previously reported the use of MALDI-TOF mass spectrometry of self-assembled monolayers (SAMs) to assay a broad range of enzyme activities (24–30). In this approach, termed SAMDI, enzyme substrates are immobilized to a SAM and then treated with solutions containing an enzyme. The monolayer is rinsed and then analyzed by SAMDI to reveal the masses of ions that correspond to the substrate-terminated alkanethiols or, as described below, biopolymers that are associated with the monolayer. Hence, when the enzyme generates a product with a mass distinct from the substrate, SAMDI can be applied for assays of the enzyme activity.
In this article, we use SAMDI mass spectrometry and oligonucleotide arrays to profile the substrate specificities of the Taq and T4 DNA ligases. We prepared arrays presenting ternary complexes of double-helical substrates that represent all possible mismatches at the 3′ side of the nick site as well as a number of deletions and insertions of bases. The arrays were treated with ligases and nucleotide triphosphate analogs and then analyzed by SAMDI mass spectrometry to determine the relative yields for both 5′-strand activation and for ligation. The resulting activity profiles reveal the relative contributions of the activation and ligation steps in determining the substrate specificities of the two ligases and point to strategies to enhance (or diminish) specificity through the use of ATP analogs.
MATERIALS AND METHODS
Materials
5-Methoxysalicyclic acid, streptavidin, cystamine–2HCl, dithiothreitol, adenosine 5′-[γ-thio]-triphosphate (ATP-γS), adenosine 5′-[β,γ-imido]-triphosphate (AMP-PNP), 10% sodium dodecyl sulfate (SDS) solution, and 20× saline-sodium citrate (SSC) buffer were obtained from Sigma-Aldrich (St. Louis, MO). Adenosine 5′-[α-thio]-triphosphate (ATP-αS) was purchased from Jena Bioscience (Jena, Germany). Ammonium citrate dibasic and glass microscope cover-slips for gold deposition were purchased from Fisher Scientific (Pittsburgh, PA). Biotin-N-hydroxysuccinimide ester was purchased from Pierce Biotechnology Inc. (Rockford, IL). Phosphate-buffered saline (PBS) was purchased from Invitrogen Corp. (Carlsbad, CA). Taq DNA ligase supplied with 10× reaction buffer (200 mM Tris–HCl, 250 mM potassium acetate, 100 mM magnesium acetate, 100 mM dithiothreitol, 10 mM NAD+ and 1% Trition X-100; pH 7.6 at 25°C) and T4 DNA ligase provided with 10× reaction buffer (500 mM Tris–HCl, 100 mM MgCl2, 100 mM DTT and 10 mM ATP; pH 7.5 at 25°C) were used as received from New England BioLabs Inc. (Ipswich, MA). Nuclease-free water was obtained from Fermentas Inc. (Hanover, MD). DNA oligonucleotides were obtained from Integrated DNA Technologies Inc. (Coralville, IA). The sequences and modifications are provided in Table 1.
Table 1.
Sequences of the nucleic acids used in this studya
| Name | Sequence |
|---|---|
| Template A (TA) | 5′-AGT AAC GGC AGA CTT CTC CTA AGG AGT CAG GTG CAC CAT G-3BioTEG-3′ |
| Template T (TT) | 5′-AGT AAC GGC AGA CTT CTC CTT AGG AGT CAG GTG CAC CAT G-3BioTEG-3′ |
| Template G (TG) | 5′-AGT AAC GGC AGA CTT CTC CTG AGG AGT CAG GTG CAC CAT G-3BioTEG-3′ |
| Template C (TC) | 5′-AGT AAC GGC AGA CTT CTC CTC AGG AGT CAG GTG CAC CAT G-3BioTEG-3′ |
| Probe 1 A (P1A) | 5′-TGC ACC TGA CTC CTA-3′ |
| Probe 1 T (P1T) | 5′-TGC ACC TGA CTC CTT-3′ |
| Probe 1 G (P1G) | 5′-TGC ACC TGA CTC CTG-3′ |
| Probe 1 C (P1C) | 5′-TGC ACC TGA CTC CTC-3′ |
| Probe 2 (P2) | 5′-5Phos-AGG AGA AGT CTG CCG-3′ |
| Probe 2 without P04 | 5′-AGG AGA AGT CTG CCG-3′ |
| Probe 1Gap1 | 5′-TGC ACC TGA CTC CT-3′ |
| Probe 1Gap2 | 5′-TGC ACC TGA CTC C-3′ |
| Probe 1lnsertA | 5′-TGC ACC TGA CTC CTG A-3′ |
| Probe 1lnsertT | 5′-TGC ACC TGA CTC CTG T-3′ |
| Probe 1lnsertG | 5′-TGC ACC TGA CTC CTG G-3′ |
| Probe 1lnsertC | 5′-TGC ACC TGA CTC CTG C-3′ |
| Internal standard20-mers | 5′-CAT GGT GCA CCT GAC TCC TG-3′ |
aThe abbreviations in the table correspond to the following modifications: 3BioTEG, a biotin modifier with a tetraethyleneglycol (TEG) spacer on the 3′-end of the DNA; 5Phos, phosphorylation of the 5′-end of the DNA. These are the same notations used by the DNA supplier (Integrated DNA Technologies, Coralville, IA, USA). The underlined bases are paired at 3′-nick site.
Immobilization of DNA to monolayers
Monolayers were prepared as previously described but with some modifications (30). Briefly, maleimide-terminated SAMs were prepared on an array of gold spots (∼4.9 mm2/spot) by immersing a gold-patterned glass cover-slip in an ethanolic solution of maleimide-terminated disulfide and tri(ethylene glycol)-terminated disulfide in a ratio of 1:9 (total concentration of disulfide was 0.5 mM) for 48 h at 4°C. Next, biotin was immobilized on the arrayed SAMs by applying a solution of N-biotinylcysteamine (50 µM in 1× PBS, pH 7.4, 30 min), and then streptavidin was immobilized by applying a solution of streptavidin (0.5 µM in 1× PBS, pH 7.4, 30 min). The monolayers were rinsed with PBS and water, dried and then treated with a solution of 3′-biotinylated oligonucleotide (1 µM in 1× PBS, pH 7.4) for 30 min at ambient temperature in a humidified chamber. The array was rinsed with buffers: 2× SSC buffer and 0.2× SSC buffer, and dried under a stream of nitrogen. Finally, probe oligonucleotides were hybridized by applying a mixture solution of probes (1 µM in 2× SSC) to the monolayers presenting the immobilized oligonucleotides for 30 min at ambient temperature. The slide was then rinsed and again dried with nitrogen.
Ligation on Chips
For ligation reactions, a ligase reaction mixture was applied to the monolayer for 1 h (unless otherwise mentioned) at 30°C in a humidified chamber. The Taq ligase reaction mixture contained Taq ligase (5 U/µl) in a ligase reaction buffer (1×: 20 mM Tris–HCl, 25 mM CH3COOK, 10 mM (CH3COO)2Mg, 10 mM DTT, 1 mM NAD+, and 0.1% Triton X-100; pH 7.6 at 25°C). Similarly, the T4 ligase mixture contained T4 DNA ligase (5 U/µl) in a ligase reaction buffer (1×: 50 mM Tris–HCl, 10 mM MgCl2, 10 mM DTT and 1 mM ATP; pH 7.5 at 25°C). Following ligation, the slide was rinsed twice with SSC buffer containing 0.2% SDS, then twice with SSC buffer, and finally twice again with SSC buffer and a solution of ammonium citrate (50 mg/ml), and dried under a stream of nitrogen.
SAMDI mass spectrometry
SAMDI mass analysis was performed using a Voyager DE-PRO Biospectrometry mass spectrometer (Applied Biosystems, Framingham, MA) with a 337 nm nitrogen laser as the desorption-ionization source. All spectra were obtained with 25 kV accelerating voltage using the linear negative mode. The grid voltage was 95% of the accelerating voltage, and the delay time was 425 ns. Approximately 1000 laser shots were integrated to generate each spectrum.
We applied a matrix solution (in a ratio of 5:1) of 5-methoxysalicyclic acid (20 mg/ml in acetonitrile) and ammonium citrate (50 mg/ml in water) onto each monolayer and then allowed the solutions to dry in air, prior to acquiring SAMDI spectra. For quantification, spectra were baseline-corrected and calibrated using the 20-mer oligonucleotide (Table 1), which is complementary to part of immobilized oligonucleotide, as an internal standard. The relative amount of each species was determined from the areas of each peak. The relative amount of adenylated intermediate and ligated product are, respectively, I/(S+I+P) and P/(S+I+P), respectively, where the peak areas were used to determine the density of substrate (S), intermediate (I) and product (P) ions.
RESULTS
Experimental design
Figure 1 illustrates the approach we used to profile the specificities of ligases. We started with SAMs of alkanethiolates on gold presenting maleimide groups (at a density of 5% relative to total alkanethiolate) against a background of tri(ethylene glycol) groups (30,31). The monolayer provides control over the density and orientation of immobilized substrates and prevents non-specific interactions between the enzymes and the surface (31–33). Furthermore, previous studies have shown that the immobilized DNA strands can adopt an orientation that makes them accessible to hybridization (34). To immobilize the template DNA strand (T), we first treated the monolayer with the biotin derivative N-biotinylcysteamine, followed by the protein streptavidin which then permitted attachment of an oligonucleotide template strand (T) that was modified with a biotin group at the 3′ terminus. The resulting monolayer was then treated with probe DNA strands (P1 and P2) to assemble the ternary complex that is a substrate for the ligases. An earlier study reported that monolayers prepared in this way were well-suited for characterization by matrix-assisted laser desorption-ionization mass spectrometry (in a technique termed SAMDI MS) (30).
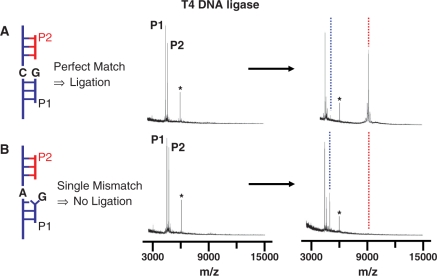
To demonstrate the assay of ligase activity, we hybridized two probes (P1G and P2, Table 1) to a template strand immobilized to the monolayer (TC, Table 1) and treated the monolayer with T4 DNA ligase (5 U/µl in reaction buffer) for 1 h at 30°C (Figure 2A). A SAMDI spectrum of the initial monolayer revealed peaks at m/z 4503.0 (calculated, m/z 4504.0) and m/z 4721.0 (calculated, m/z 4722.0), which correspond to probe 1 (P1G) and probe 2 (P2), respectively. In this and subsequent experiments, we co-immobilized a second DNA strand to the monolayer to provide an internal standard for calibrating the mass to charge values. This strand gives rise to the peak at m/z 6069.0 (Table 1). After treatment with the T4 DNA ligase, a spectrum shows new peaks at m/z 5050.7 (calculated, m/z 5051.2) and m/z 9211.0 (calculated, m/z 9208.0), which correspond to the adenylated probe 2 intermediate (blue dashed line) and ligated probe product (red dashed line), respectively. Hence, the SAMDI spectra can resolve peaks that correspond to the substrate, the adenylated probe and the ligated product. In another experiment, we immobilized probe strands that resulted in a single base pair mismatch at the nick site (A:G, TA and P1G, Table 1). Treatment of this substrate with the ligase and analysis by SAMDI revealed only one new peak at m/z 5050.1 (blue dashed line, Figure 2B), which corresponds to the adenylated probe 2. We failed to observe a peak (red dashed line) that corresponds to the ligated product. These experiments validate the use of SAMDI to observe the reaction products that result from treatment of ternary DNA complexes with ligase enzymes.
Figure 2.
Representative mass spectra showing that the SAMDI technique can identify the intermediates and products of a ligation reaction. Spectra are shown before and after the ligase reaction for a monolayer that presents (A) a substrate having a C:G matched base pair at the nick site and (B) a substrate having a A:G mismatched base pair at the nick site. In the former, the peak for the P2 probe strand (which is present in limiting quantity) gives rise to a peak representing the ligated product. For the mismatched strand, the peak for the P2 probe strand gives rise to a peak representing the adenylated probe strand, but not a ligated product. In the base pair notation X:Y, X refers to the target nucleotide and Y is the probe nucleotide. The star symbol (*) indicates the peak corresponding to the 20-mer DNA strand used for calibrating the mass range. The dotted blue and red lines indicate adenylated and ligated product, respectively.
To quantitate the extent of reaction, we baseline-corrected the mass spectra and measured the peak areas corresponding to the probe strand, the adenylated probe and the ligated product. We then determined the yield for formation of the ligated product by taking the ratio of the peak area corresponding to the ligated product to the sum of peak areas corresponding to the substrate, the adenylated intermediate and the ligated product. We find the ratios are reproducible across several independent experiments and therefore provide a meaningful relative assessment of the ligase activities. The ligation yield for the matched substrate is 82 ± 9%. We note that there appears to be an excess of the P1 strand over the P2 strand in the spectra; that is, following complete adenylation and ligation of the P2 strand, there remains a peak (though it is diminished in intensity) for the P1 strand. We believe that the ternary complex is not formed completely and therefore only a fraction of the template strand has both P1 and P2 hybridized to it. In the analysis that follows, we determine yields relative to the P2 strand and therefore are effectively excluding the presence of the binary complex (between the template and P1).
Profiling the ligation selectivity in an array format
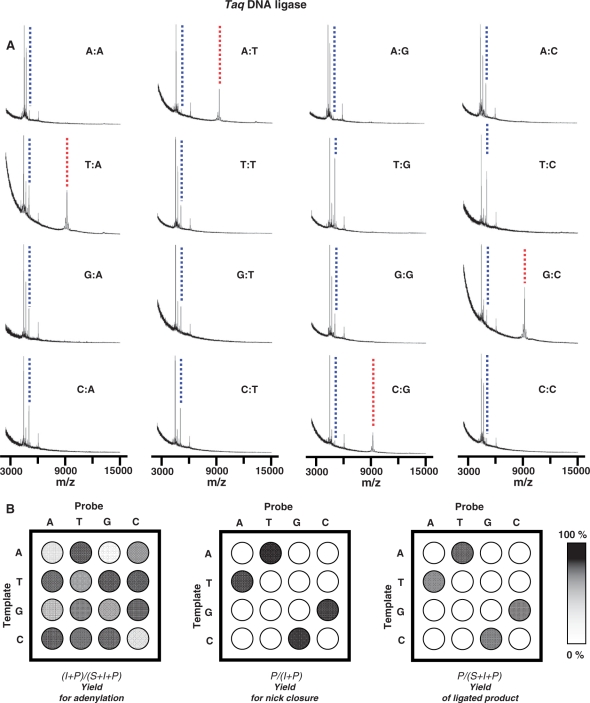
We next used an array that includes all 16 possible base pair combinations at the 3′-OH side of the nick to profile the selectivity of Taq ligase (Table 1). Identical aliquots of the ligase (5 U/µl in 1× reaction buffer) were applied to each ternary complex on the array, incubated for 1 h at 30°C, rinsed, and then analyzed by SAMDI MS. The mass spectra again reveal peaks for each of the probe strands, the adenylated intermediates and the ligated products (Figure 3A). The ligation reaction proceeded efficiently only for those substrates that had matched base pairs, which is consistent with the known selectivity of Taq DNA ligase (35). For substrates that had mismatched base pairs at the nick site, we find that the ligase often adenylated the 5′ probe strand (P2) but failed to ligate this strand with its neighbor (blue dashed line).
Figure 3.
An array was used to profile the activity of Taq DNA ligase against a panel of ternary substrates representing each possible base pair at the nick site. (A) Mass spectra reveal the extent to which the P2 probe strand was adenylated and the extent to which the two probe strands were joined. (B) The intensities of the peaks for the P2 probe strand (P2), the adenylated intermediate (I) and the ligated product (P) were used to profile the reaction. The first plot shows the yield for adenylation {(I + P)/(S + I + P)}; the second plot shows the yield for the relative activity for nick closure {P/(I + P)}; and the third plot shows the yield of ligated product {P/(S + I + P)}. The dotted blue and red lines indicate adenylated and ligated product, respectively. In the base pair notation X:Y, X refers to the target nucleotide and Y is the probe nucleotide.
We repeated this experiment three times to obtain average yields in order to determine the relative activities of the ligase for each substrate. These data are summarized in Figure 3B using a gray intensity scale to represent the yields for each step of the reaction. We first compare the yields for generating the ligated products, which are determined by taking the ratio of the peak area for ligated product to the sum of the peak areas for the probe strand, the adenylated intermediate and the ligated product {P/(S + I + P)}. This profile reveals that the two probe strands are joined only when the ternary complex has a matched base pair at the 3′ side of the junction (Figure 3B, last profile). We next compare the relative activities of the substrates for adenylation by the ligase. Because the adenylated intermediate may be converted to the ligated product, to determine the amount of adenylation, we use the ratio of the sum of peak areas of the adenylated probe and the ligated product to the sum of probe, adenylated probe and ligated product {(I + P)/(S + I + P)}. This profile reveals that the adenylation reaction is not specific for matched substrates and in fact several mismatched substrates are efficiently adenylated by the ligase (Figure 3B, first profile). Finally, we compared the relative yields for ligation of the adenylated complexes by taking the ratio of the peak area of the ligated product to the sum of the adenylated intermediate and the product {P/(I + P)}. Here we find that the specificity of Taq ligase derives primarily from this strand joining step. While several of the probe strands are efficiently adenylated, only those ternary complexes that have matched base pairs at the 3′ side of the nick are joined to give the ultimate product (Figure 3B, middle profile). These profiles also reveal a significant benefit of the SAMDI assay; by characterizing the relative activities for both steps in the ligase-mediated reaction, the basis for the specificity of the ligase is readily apparent and determined from a single experiment.
We repeated this experiment and analysis to profile the activity of T4 DNA ligase. Again, identical aliquots of the T4 ligase with ATP as the cofactor (5 U/ml in 1× reaction buffer) were applied to each ternary complex on the array, incubated for 30 min at 30°C, rinsed, and then analyzed by SAMDI MS. In contrast to the Taq ligase, T4 ligase is not specific for matched substrates (Figure 4A, last profile). We included mass spectra corresponding to the analysis in Figure 4A as we did in Figure 3 in the Supplementary Data (Figure S1). This ligase was able to ligate ternary complexes having mismatched base pairs at the nick site with the exception of substrates having A:G, G:A and C:C pairs. The profile of the relative activities of the T4 ligase for adenylation again reveals a low specificity for adenylating the 5′ strand (Figure 4A, first profile). For example, the ternary complexes having A:G, G:A and C:C mismatched base pairs were efficient substrates for adenylation even though they yielded little ligated product. We find that the second step in the reaction also exhibits poor discrimination for intermediates having matched base pairs at the nick site, but in any event is more important than the first step in determining the overall specificity (Figure 4A, middle profile).
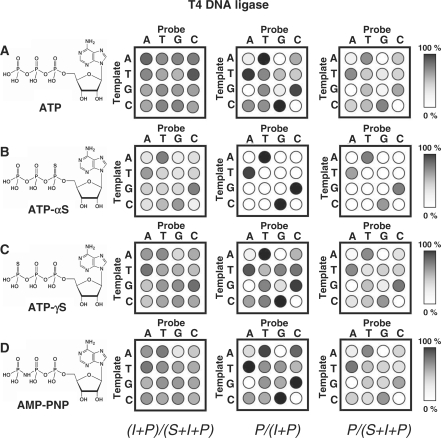
Figure 4.
An array of substrates was profiled with the T4 DNA ligase and each of four ATP analogs. Experiments were performed and analyzed as described in Figure 3. Separate experiments were performed using (A) ATP, (B) ATP-αS, (C) ATP-γS and (D) AMP-PNP as the cofactor.
We next profiled the specificity of T4 DNA ligase with three different ATP analogs: ATP-αS, ATP-γS and AMP-PNP (the structures are shown in Figure 4). Previous studies reported that T4 DNA ligases will accept several ATP analogs—including ATP-αS and AMP—though the reaction rates are slower relative to reactions that use ATP (36,37). We expected the ATP analogs to differentially affect the rates, and therefore the specificities, of the two steps in the ligase reaction. The use of ATP-αS, for example, will result in an adenylated probe strand that contains a sulfur atom that renders the diphosphate less reactive for ligation with the proximal 3′ hydroxyl group of the neighboring strand. The two other ATP analogs (ATP-γS and AMP-PNP), by contrast, may have a different reactivity in the first reaction to adenylate the probe strand but will each result in the same adenylated probe intermediate and therefore should not influence the kinetics for the strand joining step. We note that assays that omitted ATP, or its analogs, showed insignificant formation of products, revealing that the activities we observe are not due to ligases being pre-charged with the AMP group.
We first profiled T4 DNA ligase activity using each of the ATP analogs (Figure 4). With ATP-αS as the cofactor, the ligase displayed a significantly higher selectivity for ternary substrates having matched base pairs at the nick site. The ligase was still relatively non-specific in adenylating the probe strand to generate the intermediate, but showed a dramatic increase in selectivity for joining the two strands to give the ligated product. Indeed, we observed ligated products only for those substrates that have matched base pairs, and only observed products resulting from mismatched base pairs in the substrate for longer reaction times (Supplementary Figure S2). This enhanced discrimination is consistent with the lower reactivity expected for the sulfur-containing intermediate. When we repeated reactions using ATP-γS or AMP-PNP, we found that the overall selectivities do not deviate from those observed when ATP is used. There are minor changes in the relative activities of the substrates for the adenylation reaction, but no significant differences in the second step. This result is again expected, as the presence of the sulfur or nitrogen atom in the analogs may have an impact on the binding of the ATP analog to the ligase, but these analogs give rise to the ‘normal’ adenylated intermediate and therefore do not alter the relative rates by which the adenylated intermediates are converted to the ligated products.
Effect of base deletions and insertions
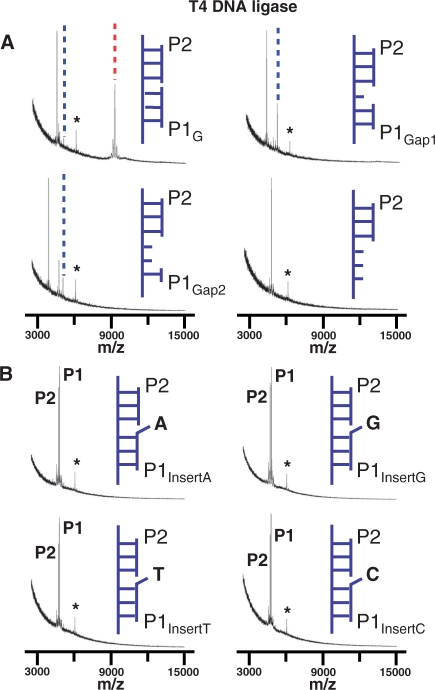
Finally, we assayed ternary complexes that introduced base deletions or insertions at the nick site of the substrate. We again immobilized a template strand to the monolayer (TC, Table 1) and then hybridized two probe DNA strands to assemble the ternary complex. We used complexes having probes with a 5′-phosphoryl group (the P2 probe) and a neighboring probe with gaps (P1Gap1 or P1Gap2, Table 1) or with insertions of each of the four nucleotides (P1InsertA, P1InsertT, P1InsertG or P1InsertC, Table 1). We applied the T4 ligase reaction cocktail (5 U/µl T4 DNA ligase in 1× reaction buffer) to each complex for 1 h at 30°C and analyzed the array with SAMDI MS. Previous work has shown that substrates with a gap of one nucleotide at a nick site can be ligated by DNA ligases including T4 DNA ligases although with substantially lower rates (38–40). The SAMDI assay revealed that the ligase could efficiently adenylate the 5′ probe strand but failed to give a ligated product (Figure 5A) for the substrate having a gap of one nucleotide (P1Gap1). We also assayed a substrate having a gap of two nucleotides (P1Gap2) and found that the ligase could still adenylate the 5′ strand, but with a lower rate than the substrate having a single deletion (Figure 5A). A binary substrate having the 5′ probe strand hybridized to the template strand but with the second probe strand omitted gave no adenylation activity, showing that both double helices are required for this step. This observation is consistent with previous reports showing that DNA ligases such as vaccinia virus DNA ligase require both the 3′ hydroxyl and the 5′ phosphoryl groups on either side of the nick for nick recognition (41,42). We assayed each of the four substrates having an insertion of the four nucleotides at the nick site and found that none of these were adenylated by the ligase enzyme. Ligation of substrates with nucleotide insertions on 3′-side of the nick has been investigated for T. kodakaraensis, A. archaeal and T. thermophilus DNA ligases, and also showed negligible ligation activities (4,43,44). It has been also reported that DNA ends with nucleotide protrusion can be ligated to blunt ends by T4 DNA ligase under certain conditions, but as expected with low efficiency (45,46).
Figure 5.
SAMDI was used to assess the relative activity of T4 DNA ligase for substrates having (A) deletions and (B) insertions of nucleotides at the nick site. In the former cases, spectra are shown for monolayers presenting a matched full length substrate, a single base deletion, a double base deletion and a substrate that does not contain the P1 probe strand. In panel B, spectra are shown for monolayers presenting substrates having an insertion of each of the four nucleotides at the nick site. The star symbol indicates the peak corresponding to the 20-mer DNA strand used for calibrating the mass range. The dotted blue and red lines indicate adenylated and ligated product, respectively.
DISCUSSION
This article describes a method that combines mass spectrometry with oligonucleotide arrays to profile the activities of DNA ligase enzymes. The use of mass spectrometry carries the benefit that it avoids the need for labeling oligonucleotides with fluorescent groups or radioisotopes. Apart from simplifying the assay format, this benefit allows independent observation of the probe strand, the intermediate adenylated strand and the ligated product. When combined with an array of substrates having all possible mismatched base pairs at the nick site, the use of mass spectrometry provided an efficient method to globally profile the specificity of the ligase for each of the two steps in the reaction. Hence, this method could clearly reveal the basis for specificity and could guide approaches to enhance (or diminish) the specificity.
One limitation of the SAMDI mass spectrometry method is that it does not directly provide quantitative measures of the reaction yields for activation and ligation of the immobilized oligonucleotides. This limitation stems from the different efficiencies with which molecules are desorbed and ionized, so that equal densities of an adenylated and non-adenylated probe strand can give peaks with different intensities. Hence, the different species on the monolayer would have to be first calibrated to identify their weighting factors in order to provide quantitative measures of reaction yields. Instead, we determine relative yields that are used to assess the specificities of the ligases. We showed that the SAMDI method provides reproducible peak intensities for samples having analytes present at a constant density. Therefore, we regard the data as ‘semi-quantitative’ in the sense that the measurements are reproducible and provide meaningful relative activities of a panel of substrates but have not been calibrated to provide absolute activities.
We used the DNA array to profile the sequence selectivity of the Taq DNA ligase. The experiment confirmed that Taq ligase requires that the substrate have a matched base pair at the 3′-hydroxyl side of the nick in order to join the probe strands (35,44). Of greater interest, the array revealed that the selectivity of the Taq DNA ligase largely owes to its stringency in the nick closure step. The ligase showed little specificity in adenylating the probe strand. Each of the matched and mismatched substrates were active in this step, though certain mismatched base pairs—including A:G, A:A, C:C and G:A—were less active in the adenylation reaction. The overall selectivity derives from the specificity of the ligase to perform the second nick closure step only for those substrates that had a matched base pair. This highly sequence-specific nature of the Taq DNA ligase underlies its use in DNA diagnostics as well as in maintaining sequence integrity in the nucleus (47–49).
We also profiled the specificity of T4 DNA ligase, which is known to ligate substrates that have mismatched base pairs at the nick site. Indeed, we confirmed that the ligase can repair the nick site for most of the substrates—excluding those having A:G, G:A and C:C base pairs—with several of the mismatched substrates displaying a high efficiency (3,38). The arrays again revealed that the specificity of this ligase derives from the second step in the reaction. Even those substrates that failed to give ligated products were actively adenylated by the enzyme. This importance of the nick closure step in controlling ligation specificity has been demonstrated for specific substrates (4,38). For example, Verly and coworkers have shown that a mismatched base pair (A:A) at the 3′ side of a nick does not hinder the adenylation reaction by T4 DNA ligases, but dramatically slows the strand joining reaction (38).
We demonstrated that the substitution of ATP with ATP-αS serves to enhance the substrate specificity of the T4 DNA ligase. This improved specificity derives from a more specific strand joining reaction, consistent with the lower electrophilicity of the thio-adenylated probe strand. Previous work has demonstrated specific examples of the effect of the ATP-αS substitution on ligase activity, but has not recognized the opportunity for enhancing the overall specificity of the enzyme (37,41,50,51). For example, one report found that yeast ligase could use ATP-αS to generate the thioadenylated intermediate, but gave poor yields for the overall reaction (50). Another example, however, showed that T4 DNA ligase can use ATP-αS, but with a 2-fold reduction in the rate of ligation (37). Most studies seeking to improve the specificity of T4 ligase identified the use of elevated temperatures and higher salt or spermidine concentrations to achieve this aim (7,52).
Finally, we also demonstrated applications of the SAMDI method to profile the effect of 3′-base deletions and insertions on T4 ligase activity. The ligase was sensitive to base deletions and insertions at the ligation junction, and did not produce ligated products. Specifically, we found that substrates having two-base deletions were still efficiently adenlyated but failed to undergo the strand-joining reaction. Previous reports have also shown that the ligation of so-called gapped substrates is slow, and can require several hours (38,53).
The primary advance reported in this paper is the use of an oligonucleotide array to profile the activity of a ligase against a panel of substrates. Importantly, the use of an array makes it straightforward to perform multiple assays in parallel and the use of mass spectrometry allows each of the intermediates in the ligase-mediated reaction to be observed. Hence, the arrays provide a global profile of the specificity of an enzyme, and for enzymes that perform multiple reactions the profiles can reveal the basis for the specificity. We also note that the SAMDI assay has the characteristics required for efficient high-throughput screening to identify activators or inhibitors of enzymes and may be used to identify reagents that modulate the activities of enzymes that act on nucleic acid substrates (29). In this way, the work extends on previous reports that have detailed specific examples of the specificity of the ligase and the role of adenylation and the strand-joining reaction. We believe that this combination will enable studies of a broader range of enzymes that act on DNA substrates.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Cao W. DNA ligases and ligase-based technologies. Clin. Appl. Immunol. Rev. 2001;2:33–43. [Google Scholar]
- 2.Cao W. DNA ligases: Structure, function and mechanism. Curr. Org. Chem. 2002;6:827–839. [Google Scholar]
- 3.Lamarche BJ, Showalter AK, Tsai M. An error-prone viral DNA ligase. Biochemistry. 2005;44:8408–8417. doi: 10.1021/bi047706g. [DOI] [PubMed] [Google Scholar]
- 4.Tong J, Cao W, Barany F. Biochemical properties of a high fidelity DNA ligase from thermus species AK16D. Nucleic Acids Res. 1999;27:788–794. doi: 10.1093/nar/27.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 6.Bullard DR, Bowater RP. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu DY, Wallace RB. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene. 1989;76:245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- 8.Sriskanda V, Shuman S. Chlorella virus DNA ligase: nick recognition and mutational analysis. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kletzin A. Molecular characterization of a DNA ligase gene of the extremely thermophilic archaeon Desulfurolobus ambivalens shows close phylogenetic relationship to eukaryotic ligases. Nucleic Acids Res. 1992;20:5389–5396. doi: 10.1093/nar/20.20.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos W, Tappe N, Talamantez J, Friedberg EC, Tomkinson AE. Two distinct DNA ligase activities in mitotic extracts of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:1485–1492. doi: 10.1093/nar/25.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhagwat AS, Sanderson RJ, Lindahl T. Delayed DNA joining at 3′ mismatches by human DNA ligases. Nucleic Acids Res. 1999;27:4028–4033. doi: 10.1093/nar/27.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Marjory AS, Keller M, Aujay M, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Subramanya HS, Doherty AJ, Ashford SR, Wigley DB. Crystal structure of an ATP-dependent DNA ligase from bacteriophase T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 14.Tomkinson AE, Vijayakumar S, Pascal JW, Ellenberger T. DNA ligases: Structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 15.Shuman S. DNA ligases: progress and prospects. J. Biol. Chem. 2009;284:17365–17369. doi: 10.1074/jbc.R900017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott BOS, Lavesa-Curto M, Bullard DR, Butt JN, Bowater RP. Immobilized DNA hairpins for assay of sequential breaking and joining of DNA backbones. Anal. Biochem. 2006;358:90–98. doi: 10.1016/j.ab.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Gul S, Brown R, May E, Mazzulla M, Smyth MG, Berry C, Morby A, Powell DJ. Staphylococcus aureus DNA ligase: characterization of its kinetics of catalysis and development of a high-throughput screening compatible chemiluminescent hybridization protection assay. Biochem. J. 2004;383:551–559. doi: 10.1042/BJ20040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by mass spectrometry. Mass Spectrom. Rev. 2002;21:388–418. doi: 10.1002/mas.1009. [DOI] [PubMed] [Google Scholar]
- 19.Stoerker J, Mayo JD, Tetzlaff CN, Sarracino DA, Schwope I, Richert C. Rapid genotyping by MALDI-monitored nuclease selection from probe libraries. Nat. Biotechnol. 2000;18:1213–1216. doi: 10.1038/81226. [DOI] [PubMed] [Google Scholar]
- 20.Koster H, Tang K, Fu D, Braun A, van den Boom D, Smith CL, Cotter RJ, Cantor CR. A strategy for rapid and efficient DNA sequencing by mass spectrometry. Nat. Biotechnol. 1996;14:1123–1128. doi: 10.1038/nbt0996-1123. [DOI] [PubMed] [Google Scholar]
- 21.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tost J, Schatz P, Schuster M, Berlin K, Gut IG. Analysis and accurate analysis of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31:e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonk T, Humeny A, Gebert J, Sutter C, von Knebel Doeberitz M, Becker CM. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based detection of microsatellite instabilities in coding DNA sequences: a novel approach to identify DNA-mismatch repair-deficient cancer cells. Clin. Chem. 2003;49:552–561. doi: 10.1373/49.4.552. [DOI] [PubMed] [Google Scholar]
- 24.Gurard-Levin ZA, Mrksich M. Combining self-assembled monolayers and mass spectrometry for applications in biochips. Annu. Rev. Anal. Chem. 2008;1:767–800. doi: 10.1146/annurev.anchem.1.031207.112903. [DOI] [PubMed] [Google Scholar]
- 25.Min D, Su J, Mrksich M. Profiling kinase activities by using a peptide chip and mass spectrometry. Angew. Chem. Int. Ed. 2004;43:5973–5977. doi: 10.1002/anie.200461061. [DOI] [PubMed] [Google Scholar]
- 26.Su J, Rajapaksha TW, Peter ME, Mrksich M. Assays of endogenous caspase activities: a comparison of mass spectrometry and fluorescence formats. Anal. Chem. 2006;78:4945–4951. doi: 10.1021/ac051974i. [DOI] [PubMed] [Google Scholar]
- 27.Min D, Yeo W, Mrksich M. A method for connecting solution-phase enzyme activity assays with immobilized format analysis by mass spectrometry. Anal. Chem. 2004;76:3923–3929. doi: 10.1021/ac049816z. [DOI] [PubMed] [Google Scholar]
- 28.Houseman BT, Mrksich M. The role of ligand density in the enzymatic glycosylation of carbohydrates presented on self-assembled monolayers of alkanethiolates on gold. Angew. Chem. Int. Ed. 1999;38:782–785. doi: 10.1002/(SICI)1521-3773(19990315)38:6<782::AID-ANIE782>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 29.Min D, Tang W, Mrksich M. Chemical screening by mass spectrometry to identify inhibitors of anthrax lethal factor. Nat. Biotechnol. 2004;22:717–723. doi: 10.1038/nbt973. [DOI] [PubMed] [Google Scholar]
- 30.Tsubery H, Mrksich M. Biochemical assays of immobilized oligonucleotides with mass spectrometry. Langmuir. 2008;24:5433–5438. doi: 10.1021/la7040482. [DOI] [PubMed] [Google Scholar]
- 31.Houseman BT, Gawalt ES, Mrksich M. Maleimide-functionalized self-assembled monolayers for the preparation of peptide and carbohydrate biochips. Langmuir. 2003;19:1522–1531. [Google Scholar]
- 32.Mrksich M, Whitesides GM. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Annu. Rev. Biophys. Biomol. Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 33.Houseman BT, Mrksich M. Towards quantitative assays with peptide chips: a surface engineering approach. Trends Biotechnol. 2002;20:279–281. doi: 10.1016/s0167-7799(02)01984-4. [DOI] [PubMed] [Google Scholar]
- 34.Lee C, Gong P, Harbers GM, Grainger DW, Castner DG, Gamble LJ. Surface coverage and structure of mixed DNA/alkylthiol monolayers on gold: Characterization by XPS, NEXAFS, and Fluorescence intensity measurements. Anal. Chem. 2006;78:3316–3325. doi: 10.1021/ac052137j. [DOI] [PubMed] [Google Scholar]
- 35.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raae AJ, Kleppe K. Effect of ATP analogues on T4 polynucleotide ligase. Biochem. Biophys. Res. Commun. 1978;81:24–27. doi: 10.1016/0006-291x(78)91625-x. [DOI] [PubMed] [Google Scholar]
- 37.Montecucco A, Lestingi M, Pedrali-noy G, Spadari S, Ciarrocchi G. Use of ATP, dATP and their α-thio derivatives to study DNA ligase adenylation. Biochem. J. 1990;271:265–268. doi: 10.1042/bj2710265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goffin C, Bailly V, Verly WG. Nicks 3′ or 5′ to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucleic Acids Res. 1987;15:8755–8771. doi: 10.1093/nar/15.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho CK, Van Etten JL, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by Chlorella virus PBCV-1. J. Virol. 1997;71:1931–1937. doi: 10.1128/jvi.71.3.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng C, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by haemophilus influenzae. Nucleic Acids Res. 1997;25:1369–1374. doi: 10.1093/nar/25.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuman S. Vaccinia virus DNA ligase: Specificity, fidelity, and inhibition. Biochemistry. 1995;34:16138–16147. doi: 10.1021/bi00049a029. [DOI] [PubMed] [Google Scholar]
- 42.Sekiguchi J, Shuman S. Nick sensing by vaccinia virus DNA ligase requires a 5′ phosphate at the nick and occupancy of the adenylate binding site on the enzyme. J. Virol. 1997;71:9679–9684. doi: 10.1128/jvi.71.12.9679-9684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatani M, Ezaki S, Atomi H, Imanaka T. Substrate recognition and fidelity of strand joining by an archaeal DNA ligase. Eur. J. Biochem. 2002;269:650–656. doi: 10.1046/j.0014-2956.2001.02695.x. [DOI] [PubMed] [Google Scholar]
- 44.Tong J, Barany F, Cao W. Ligation reaction specificities of NAD+-dependent DNA ligase from the hyperthermophile Aquifex aeolicus. Nucleic Acids Res. 2000;28:1447–1454. doi: 10.1093/nar/28.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cimmino C, Santori F, Donini P. Ligation of nonmatching DNA molecule ends. Plasmid. 1995;34:1–10. doi: 10.1006/plas.1995.1028. [DOI] [PubMed] [Google Scholar]
- 46.Wiaderkiewicz R, Ruiz-Carrillo A. Mismatch and blunt to protruding-end joining by DNA ligases. Nucleic Acids Res. 1987;15:7831–7847. doi: 10.1093/nar/15.19.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Chu X, Liu Y, Jiang J, He Z, Zhang Z, Shen G, Yu R. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol. 2000;18:561–564. doi: 10.1038/75452. [DOI] [PubMed] [Google Scholar]
- 49.Baron H, Fung S, Aydin A, Bahring S, Luft FC, Schuster H. Oligonucleotide ligation assay (OLA) for the diagnosis of familial hypercholesterolemia. Nat. Biotechnol. 1996;14:1279–1282. doi: 10.1038/nbt1096-1279. [DOI] [PubMed] [Google Scholar]
- 50.Tomkinson AE, Tappe NJ, Friedberg EC. DNA ligase I from Saccharomyces cerevisiae: Physical and Biochemical characterization of the CDC9 gene product. Biochemistry. 1992;31:11762–11771. doi: 10.1021/bi00162a013. [DOI] [PubMed] [Google Scholar]
- 51.Elder RH, Rossignol J. DNA ligases from rat liver. Purification and partial characterization of two molecular forms. Biochemistry. 1990;29:6009–6017. doi: 10.1021/bi00477a019. [DOI] [PubMed] [Google Scholar]
- 52.Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson SV, Magnusson G. Sealing of gaps in duplex DNA by T4 DNA ligase. Nucleic Acids Res. 1982;10:1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.