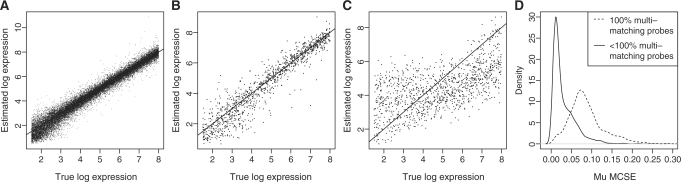
Figure 4.
Plots showing the ability of MMBGX to recover gene-level log expression values from simulated human Gene array data. (A) The scatterplot shows that the model implementation recovers the simulated expression values, μgc, for non-multi-mapping probesets accurately; (B) the scatterplot shows that the expression for multi-mapping probesets with one or more probes that uniquely match the intended transcript is also well-estimated; (C) the scatterplot shows that the expression for multi-mapping probesets with no probes that uniquely match the intended transcript has higher variance and shrinkage towards the mean at low and high levels of expression, μgc; (D) the density lines show that the shrunk estimates of μgc (i.e. those hailing from multi-mapping probesets with no single-match probes) have a higher MCSE than the non-shrunk estimates (i.e. hailing from multi-mapping probesets with at least one single-match probe).