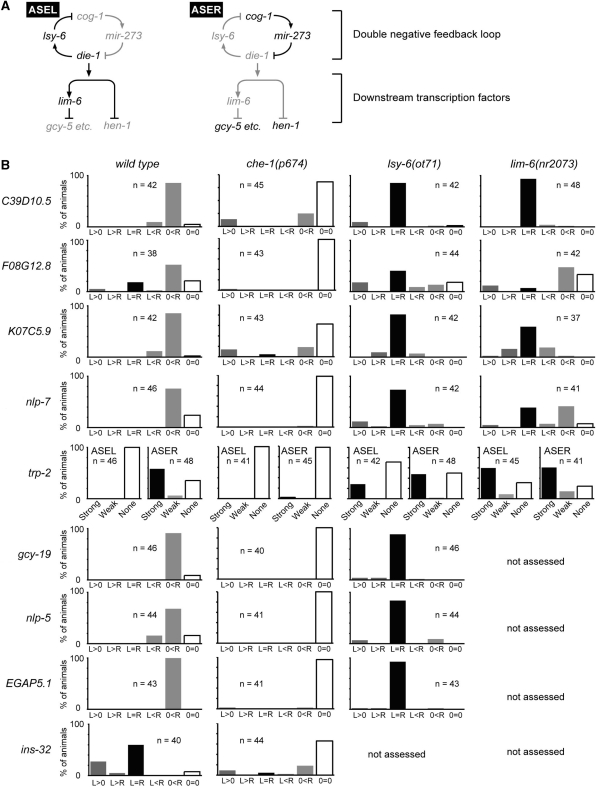
Figure 3.
Quantification of the expression bias of the newly identified genes in wild-type and mutant backgrounds. (A) Lateralized gene expression in ASE is controlled by the upstream double-negative feedback loop and the downstream transcription factors (6). ASER-biased gcy genes are regulated by lim-6, whereas hen-1 is independent of lim-6. Genes in active or inactive state are shown in black or grey, respectively. (B) Quantification of the expression laterality in ASE neurons of the newly identified genes in wild-type and the mutant backgrounds. The expression bias is indicated as follows except for trp-2: ‘L > 0’ and ‘0 < R’ refer to the restricted expression to ASEL or ASER, respectively; ‘L > R’ and ‘L < R’ refer to different levels of expression in ASEL versus ASER; ‘L = R’ refers to equal expression in ASEL and ASER; ‘0 = 0’ refers to no expression in ASEL nor in ASER. For trp-2, since strong Venus expression in trp-2p::Venus worms were seen in neurons just medial to ASE, the expression was observed on one side for each animal. ‘Strong’, ‘Weak’ and ‘None’ refer to the expression levels at each neuron. The number of examined animals is shown in each graph.