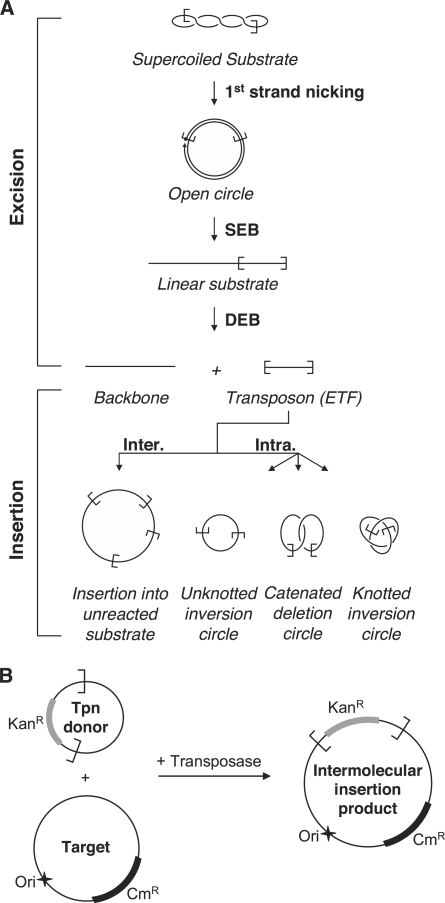
Figure 1.
In vitro Hsmar1 transposition assays. (A) A schematic representation of the different steps of a mariner transposition reaction using a supercoiled plasmid as the transposon donor. First strand nicking at one transposon end generates an open circular product in which the 5′-end of the transposon is separated from the donor sequence. Second strand nicking exposes the 3′-OH at the transposon end and linearizes the donor yielding the SEB product. A similar sequence of nicks at the other transposon end yields the DEB products, which are the plasmid backbone plus the excised transposon fragment (ETF). Excision is followed by insertion of the transposon in one of the various possible targets. Examples of inter- and intramolecular integration events are shown [for further details of these products see Chalmers and Kleckner (26)]. (B) The ‘in vitro hop’ assay for the quantification of intermolecular integration events. The transposon donor encodes a kanamycin resistance marker (KanR) flanked by Hsmar1 transposon ends. In vitro transposition is performed in presence of a target plasmid encoding a chloramphenicol resistance marker (CmR). The intermolecular transposition efficiency was obtained by transformation in E. coli and by dividing the number of colonies obtained after selection on Kan+Cm by the number of colonies obtained on Cm alone. No colonies were recovered if the donor, the target or the transposase were omitted. The donor plasmid has a conditional origin of replication that does not function in the recipient strain. This serves to eliminate any bias introduced by double transformation events in which a cell receives copies of both the donor and the target plasmid.