Abstract
HIV-1 nonsubtype B variants account for the majority of HIV infections worldwide. Drug resistance in individuals who have never undergone antiretroviral therapy can lead to early failure and limited treatment options and, therefore, is an important concern. Evaluation of reported transmitted drug resistance (TDR) is challenging owing to varying definitions and study designs, and is further complicated by HIV-1 subtype diversity. In this article, we discuss the importance of various mutation lists for TDR definition, summarize TDR in nonsubtype B HIV-1 and highlight TDR reporting and interpreting challenges in the context of HIV-1 diversity. When examined carefully, TDR in HIV-1 non-B protease and reverse transcriptase is still relatively low in most regions. Whether it will increase with time and therapy access, as observed in subtype-B-predominant regions, remains to be determined.
Keywords: diversity, drug resistance, HIV-1, nonsubtype B, resistance transmission, transmitted drug resistance, treatment-naive
The use of antiretroviral therapy has resulted in the emergence of HIV drug resistance [1–7]. In resource-rich settings, such as North America, Europe and Australia, significant resistance has been observed to the three main antiretroviral drug classes, nucleoside reverse-transcriptase (RT) inhibitors (NRTIs), non-NRTIs (NNRTIs) and protease inhibitors (PIs) [8–14]. Primary, or transmitted drug resistance (TDR), is defined as resistance to one or more antiretroviral drugs found in individuals with no previous drug exposure and is attributed to the direct transmission of resistant strains from treated individuals. As access to HAART is rolled out globally [15,16], the monitoring of TDR is essential, especially in areas where availability of second-line drugs is limited and the selection of first-line drug regimens is vital for effective treatment [17–19].
Evaluation of TDR is important in order to assess the efficacy of drug regimens, to determine optimal treatment regimens for a particular region and to measure the impact of risk-modifying interventions on HIV transmission. Current guidelines in resource-rich settings suggest resistance testing at initiation of care, regardless of whether HAART is planned or whether the infection occurred recently [9,20–22]. In resource-limited settings, the WHO recommends surveillance of TDR as a comprehensive approach to HIV care [23,24].
Several factors contribute to the reporting and the occurrence of TDR in a given population. Study methodology, such as the specific drug-resistance mutation list used to interpret resistance data, can greatly affect the reported prevalence of transmitted resistance and, until recently, there has been no consensus on which resistance mutations should be classified as markers of TDR. Other factors that can affect TDR occurrence include the number of patients receiving antiretroviral treatment in the population, their risky behaviors (e.g., unprotected sex and intravenous drug use), nonadherence to treatment, current or previous HAART regimens efficacy, available treatment-monitoring strategies, rates of virologic suppression and viral fitness of drug-resistant variants [25]. In areas where HAART has been available since its development, such as North America and Europe, TDR has been reported to be as high as 25% [12,13,26,27].
The detection of TDR is dependent on various factors, such as duration of infection (mutations may disappear with time [28]) and sensitivity of testing methods (current common assays only detect mutations present in >15–30% of the viral quasispecies). During the course of HIV infection, certain viral populations may predominate according to widespread sequencing methods, but resistant minor variants often exist [29–36] and lead to earlier treatment failure [3,32,37,38]. The identification of TDR is further complicated by the diversity of HIV across different types (HIV-1 and -2), groups (main [M], outlier [O] and new [N]), subtypes and recombinant forms [7,39–42] that are prevalent worldwide [43]. Group M accounts for over 99% of globally reported HIV/AIDS infections and is further classified into nine pure subtypes (A, B, C, D, F, G, H, J and K) and many circulating and other recombinant forms [301]. Nonsubtype B infections account for approximately 90% of HIV infection worldwide, but are the least studied owing to financial and infrastructure limiations in geographical regions where nonsubtype B predominates. Subtype B is predominant in resource-rich settings where access to HAART has been widely available for over 10 years. In this article, we summarize reported data on PI, NRTI and NNRTI TDR in nonsubtype B HIV-1 and discuss its relevance and challenges.
Definition of TDR
Major challenges in the evaluation of TDR include the extensive diversity of HIV protease and RT genes [40], the large number of HIV-associated drug-resistance mutations [44] and the lack of a standard definition of which mutations are linked with antiretroviral treatment failure. Certain mutations known to be associated with drug resistance in subtype B are naturally occurring mutations (polymorphisms) in other subtypes and, therefore, should be excluded as markers of TDR [45]. Although some of these mutations may indeed be transmitted, including polymorphisms as TDR mutations will lead to artificially inflated rates of TDR.
Various lists of drug-resistance mutations are used in different interpretation algorithms (Table 1), such as those from the International AIDS Society-USA (IAS-USA [44]), The French National Agency for AIDS Research (ANRS [302]), The Stanford University HIV Drug Resistance Database [303] and The Rega Institute [304]. These are comprehensive lists of amino acid mutations that are associated with drug resistance, derived mostly from research in subtype-B HIV-1-infected individuals. These lists include mutations that were identified by in vitro experiments, drug susceptibility testing, clinical experience in patients failing therapy and genotypic studies. Inclusively, these four lists contain 100 mutations in 37 RT positions and 118 mutations in 46 protease positions that are associated with drug resistance. Although the lists seem similar, there are only 38 positions (12 NRTI, 11 NNRTI and 15 PI related; bold in Table 1) in which mutations appear in all four lists, emphasizing the importance of putting reported data in the context of which list is used, as well as potential interpretation discordances [46–49]. These methods may overestimate TDR prevalence because they are a compilation of all known drug-resistant mutations, including many that occur as natural polymorphisms in nonsubtype B viruses.
Table 1. HIV-1 drug-resistance mutation lists.
| ANRS | IAS | REGA | STAN | SDRM | ||
|---|---|---|---|---|---|---|
| Nucleoside reverse-transcriptase inhibitors mutations* | ||||||
| M41 | L | L | L | L | L | |
| E44 | D | – | A/D | – | – | |
| A62 | – | V | – | – | – | |
| K65 | R | R | N/R | N/R | R | |
| D67 | N | N | G/N/del | N | E/N/G | |
| T69 | D/N/S/i | i | A/N/D/G/i | i | D/i | |
| K70 | R | E/R | E/G/R | E/G/R | E/R | |
| L74 | I/V | V | I/V | I/V | I/V | |
| V75 | A/M/S/T | I | A/I/M/S/T | I/M/T | A/M/S/T | |
| F77 | – | L | L | L | L | |
| Y115 | F | F | F | F | F | |
| F116 | – | Y | Y | Y | Y | |
| V118 | – | – | I | – | – | |
| Q151 | M | M | M | M | M | |
| M184 | I/V | I/V | I/V | I/V | I/V | |
| L210 | W | W | W | W | W | |
| T215 | Mul‡ | F/Y | Mul‡ | F/Y | C/D/E/F/I/S/V/Y | |
| K219 | E/Q | E/Q | E/H/N/R/Q | E/Q | E/N/R/Q | |
| Non-nucleoside reverse-transcriptase inhibitors mutations* | ||||||
| V90 | I | I | I | – | – | |
| A98 | G/S§ | G | G | G | – | |
| L100 | I | I | I | I | I | |
| K101 | E/P | E/H/P | E/H/N/P/Q | E/P | E/P | |
| K103 | H/N/S/T | N | H/N/R/S/T | N/S | N/S | |
| V106 | A/I/M | A/I/M | A/I/M | A/M | A/M | |
| V108 | – | I | I | I | – | |
| E138 | – | A | K/Q | – | – | |
| V179 | D/F | D/F/T | D/E | D/E/F | F | |
| Y181 | C/I/V | C/I/V | C/I/V | C/I/V | C/I/V | |
| Y188 | C/L/H | C/L/H | C/L/H | C/L/H | C/L/H | |
| G190 | Mul¶ | A/S | Mul¶ | A/E/S | A/E/S | |
| H221 | – | – | Y | – | – | |
| P225 | H | H | H | H | H | |
| F227 | – | – | C/L | C/L | – | |
| M230 | L | L | I/L | L | L | |
| P236 | – | – | L | L | – | |
| K238 | – | – | N/T | T | – | |
| Y318 | – | – | F | – | – | |
| Protease inhibitors mutations* | ||||||
| L10 | F/I/M/R/V | C/F/I/R/V | I/F/V/Y | – | – | |
| V11 | I | I | I/L | – | – | |
| I13 | – | V | – | – | – | |
| I15 | A/V | – | – | – | – | |
| G16 | E | E | – | – | – | |
| K20 | I/M/R/T | I/M/R/T/V | I/M/R/T/V | – | – | |
| L23 | – | – | I | I | I | |
| L24 | I | I | F/I | I | I | |
| D30 | N | N | N | N | N | |
| V32 | I | I | I/L | I | I | |
| L33 | F/I/V | F/I/V | F/I/M/V | F | – | |
| E34 | – | Q | V | – | – | |
| E35 | – | G | G/N | – | – | |
| M36 | I/L/V | I/L/V | – | – | – | |
| L38 | – | – | W | – | – | |
| R41 | – | – | I/T | – | – | |
| K43 | – | T | R/T | – | – | |
| K45 | – | – | I | – | – | |
| M46 | I/L | I/L | I/L | I/L | I/L | |
| I47 | A/V | A/V | A/V | A/V | A/V | |
| G48 | V | V | A/M/V | M/V | M/V | |
| I50 | L/V | L/V | L/V | L/V | L/V | |
| F53 | L/W/Y | L/Y | L | L | L/Y | |
| I54 | A/L/M/S/T/V | A/L/M/S/T/V | A/C/L/M/S/T/V | A/L/M/T/V | A/L/M/S/T/V | |
| Q58 | E | E | E | - | – | |
| D60 | E | E | – | – | – | |
| I62 | V | V | V | – | – | |
| L63 | P | P | – | – | – | |
| I64 | – | L/M/V | M/V | – | – | |
| I66 | – | – | F | – | – | |
| H69 | I/K/N/Q/R/Y | K | – | – | – | |
| K70 | – | – | E | – | – | |
| A71 | I/L/T/V | I/L/T/V | I/L/T/V | – | – | |
| G73 | A/S/T | A/C/S/T | A/C/F/S/T/V | S/T | A/C/S/T | |
| T74 | P | P | A/E/S/P | – | – | |
| L76 | V | V | V | V | V | |
| V77 | I | I | A/T/V | – | – | |
| V82 | A/C/G/F/M/S/T | A/F/L/I/S/T | A/F/L/M/S/T | A/F/L/S/T | A/C/F/L/M/S/T | |
| N83 | – | D | D | – | D | |
| I84 | A/V | V | A/C/V | A/C/V | A/C/V | |
| I85 | V | V | V | – | V | |
| N88 | D/S | D/S | D/S | D/S | D/S | |
| L89 | I/M/R/T/V | V | I/T/V | – | – | |
| L90 | M | M | M | M | M | |
| I93 | – | L/M | M | – | – | |
| C95 | – | – | F | - | - | |
Positions in bold have mutations that appear in all five lists.
Wild-type amino acid (for consensus subtype B sequence) and reverse transcriptase or protease position.
ANRS: T215A/C/D/E/F/G/H/I/L/N/S/V/Y.
For subtype C only.
ANRS: G190A/C/E/Q/S/T/V.
–: Mutation not on list; A: Alanine; ANRS: The French National Agency for AIDS Research, v17 [302]; C: Cysteine; D: Aspartic acid; del: Deletion; E: Glutamic acid; F: Phenylalanine; G: Glycine; H: Histidine; i: Insertion; I: Isoleucine; IAS: International AIDS Society–USA; K: Lysine; L: Leucine; M: Methionine; Mul: Multiple mutations; N: Asparagine; P: Proline; Q: Glutamine; R: Arginine; REGA: Rega Institute mutation list [304]; S: Serine; SDRM: Surveillance drug-resistance mutation list; STAN: Stanford University HIV Drug Resistance Database [303]; T: Threonine; V: Valine; W: Tryptophan; Y: Tyrosine.
In an effort to increase the accuracy of identifying TDR mutations, Bennett et al. published the original and recently updated surveillance drug-resistance mutation (SDRM) list [50,51], compiled as a more specific method to distinguish between polymorphic variation and ‘real’ TDR mutations (Table 1). There are several noticeable differences between the SDRM list and the ‘regular’ lists. For example, M36I (methionine converted to isoleucine at protease position 36) is an accessory resistance mutation that was reported to increase viral fitness and confer resistance to ritonavir and nelfinavir [44,52]. This position is extremely variable in nonsubtype B viruses [40,45,53,54] where isoleucine is the wild-type consensus amino acid at this position in subtypes A, C, D, F and G and circulating recombinant form (CRF)01_AE. This mutation, which is present on the IAS-USA and ANRS mutation lists, is excluded from the SDRM list, as are a total of 79 protease and 47 RT mutations that are absent from the SDRM list but present on other lists. These exclusions allow for a more accurate, not inflated, estimation of TDR. Although a recent report from the UK suggests this may be negligible [55], in smaller populations, such as those that are recommended by the WHO resistance-surveillance protocols, as well as in areas where non-B subtypes predominate, the effect could be magnified and may lead to inaccurate overestimation of TDR.
Epidemiology & prevalence of TDR in nonsubtype B HIV-1
It is difficult to adequately estimate the prevalence of TDR in non-B HIV-1 subtypes from the published literature, mainly owing to variation in study design and lack of standard definitions and classifications. Despite these limitations, most studies do report mutations in untreated individuals who are infected with non-B subtypes, which appear on resistance mutation lists and confer high-level resistance to one or more antiretroviral drugs. In resource-rich settings, where HIV-1 subtype B predominates, TDR rates as high as 16–25% in the USA [27,56,57] and 9–14% in western Europe [12,13,58–60] are measured. This is likely due to an early and gradual introduction of antiretroviral therapy in these populations.
Studies from resource-limited settings, where HIV-1 non-B subtypes predominate, usually report TDR from small populations in limited epidemiological samples. We aggregated data and reviewed 130 studies in the published literature that examined drug resistance in sequences from 9984 individuals infected with nonsubtype B HIV-1 (subtypes A: 1755; C: 2423; D: 447; F: 145; G: 420; H/J/K: 65; CRF01_AE: 1238; CRF02_AG: 1243; other recombinant forms: 1813; and unknown nonsubtype B strains that were not classified: 435). We reviewed studies that focus on, or include, more than five nonsubtype B infected, untreated, individuals that possessed resistance mutations; studies in which it was unclear whether HIV-infected patients received antiretroviral therapy were excluded. Studies that did not specifically comment on resistance mutations, but whose sequences are available in the Stanford University HIV Drug Resistance Database were also included. We compiled drug-resistance mutations as reported in the studies and included major mutations according to the IAS-USA list, which was used by many studies, and when possible, added TDR mutations based on the SDRM list. Only four (RT A62V, V75I, V108I and PR L33F) of 52 major resistance mutations according to the IAS-USA list are not considered markers of TDR per the SDRM list.
Transmitted drug resistance in nonsubtype B HIV-1 was reported from 63 countries across all continents (Africa: 25 countries; Asia: 19; Europe: 15; South America: two; Central America: one; and North America: one; Figure 1). The map in Figure 1 represents the majority of subtypes and recombinants from different geographic regions that were reported in these studies and mostly agrees with previous reports [41,43]. Notable findings include:
Figure 1. HIV-1 global diversity as reported in studies included in this article.
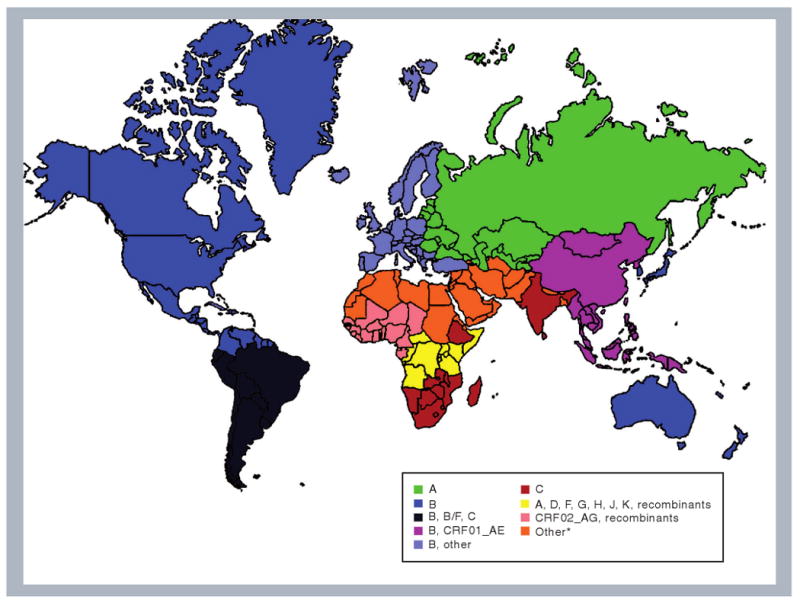
Data are based on references cited in Table 2, as well as others with subtype B data [41,141–150]. Colors indicate predominate subtype(s) in a given region based on those studies and do not necessarily include all subtypes epidemiologically reported from that region. Subtypes referred to as ‘other’ indicate numerous recombinant forms and/or a low prevalence of other subtypes. *Reports of subtypes from these regions are limited. Current data suggest existence of many different subtypes and recombinant forms with no specific predominating species. CRF: Circulating recombinant form.
A rise in reported subtype C in South America, especially Brazil;
An increase in non-B subtypes in Europe, likely secondary to immigration from African and other countries [61–68];
The existence of all major subtypes and numerous recombinant forms in Africa.
Country-specific subtype distribution is difficult to quantify secondary to sparse and varying epidemiological data, and diverse and, at times, unclear subtype identification methodologies.
Drug resistance in individuals infected with non-B HIV-1 subtypes was reported in 80 of the 130 studies (Table 2). In Africa, TDR was variable and 27 of 59 studies reported no resistance. Studies from South Africa reported increasing TDR with time, with almost no resistance in 2000–2001 [69–71] and high levels (21%) of resistance to NNRTIs in 2008, including major mutations K103N and V106M [72]. This high TDR rate was reported in a small cohort (n = 14) and may not accurately reflect actual TDR prevalence. A study from Burundi reported drug resistance of 94% to PI's in treatment-naive individuals [73]. However, this high rate is based on the prevalence of the protease M36I polymorphism, which should not be on the TDR list, as discussed previously. Additional studies that included polymorphisms as resistance mutations reported high TDR rates. The most commonly observed protease positions with high polymorphism rates were 10, 20, 36, 63, 69, 77 and 93, and rates as high as 100% were reported. Overall, TDR reported from multiple African regions was higher for RT inhibitors, probably reflecting the earlier introduction of these drugs and their inclusion in first-line HAART regimens.
Table 2. Studies of HIV reverse transcriptase and protease drug resistance in nonsubtype B-infected treatment-naive patients.
| Country | Sample years | Patients (genotypes) | Reported TDR (%; specific mutations) per drug class* | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | PI | ||||||||||
| Africa | ||||||||||||
| Africa | 1995–1999 | 142 (129) | None | 3.9 (V108I, Y181C) | 1.6 (L33F, M46I) | [151] | ||||||
| Africa | 1996–2003 | 35 (33) | None | 12.1 (V108I) | 9.1 (M46I) | [152] | ||||||
| Botswana | 2001 | 71 (71) | None | None | None | [153] | ||||||
| Burkina Faso | 2006‡ | 39 (17) | None | None | None | [154] | ||||||
| Burkina Faso | 2003–2004 | 43 (38) | None | 5.3 (K103N) | 7.9 (N88D) | [155] | ||||||
| Burkino Faso | 2001–2003 | 97 (97) | None | 4.1 (106A, V108I) | 4.1 (L33F, M46I/L) | [156] | ||||||
| Burundi | 2000 | 18 (18) | None | None | None | [73] | ||||||
| Burundi | 2002 | 119 (119) | None | 0.8 (G190E) | None | [157] | ||||||
| Cameroon | 1998 | 110 (109) | Not reported | Not reported | 0.9 (L24I) | [119] | ||||||
| Cameroon | 2004 | 54 (54) | 3.7 (V75I, M184V) | 3.7 (Y188C, L100I) | 7.4 (M46I/L, V82A) | [158] | ||||||
| Cameroon | 2000–2002 | 128 (128) | Not studied | Not studied | None | [126] | ||||||
| Cameroon | 2007‡ | 40 (24§) | 8.3§ (D67E, M184V) | 4.2§ (Y188H) | 8.3§ (L33F, M46I) | [159] | ||||||
| Cameroon | 2004 | 79 (78) | 3.8 (T215Y/F) | 9.0 (L100I, V108I, Y181C, L 210W) | 2.6 (M46L) | [160] | ||||||
| Cameroon | 1999 | 75 (70§) | Not reported | Not reported | 4.3§ (L33F, G73S) | [161] | ||||||
| Cameroon | 1999 | 47 (19) | None | None | 5.3 (V108I) | [162] | ||||||
| Cameroon | 2001–2003 | 102 (102) | 2.0 (A62V, M184V) | 1.0 (V108I) | 2.9 (L33F, M46I/L) | [156] | ||||||
| Cameroon | 2001–2004 | 110 (96) | 1.0 (L210W) | None | 1.0 (N88S) | [163] | ||||||
| CAR | 2005 | 150 (114) | None | None | None | [164] | ||||||
| CAR | 2002 | 38 (12) | Not studied | Not studied | None | [165] | ||||||
| Cote d'Ivoire | 2003‡ | 20 (20) | None | None | None | [113] | ||||||
| Cote d'Ivoire | 1997–2000 | 99 (99) | None | None | None | [129] | ||||||
| Cote d'Ivoire | 2001–2002 | 107 (107) | 0.9 (K219Q) | 3.1 (K101E, K103N) | 2.0 (F53Y, N88D) | [166] | ||||||
| DRC | 2002 | 70 (70) | None | 1.4 (K103N) | 2.9 (L90M, M46L) | [167] | ||||||
| Ethiopia | 2003 | 92 (92) | 1.1 (V75I) | 2.2 (G190A) | None | [124] | ||||||
| Ethiopia, Botswana | 1994–1995 | 14 (14) | Not reported | Not reported | None | [168] | ||||||
| Gabon | 2007 | 25 (22) | None | None | None | [169] | ||||||
| Gabon | 1996–1999 | 41 (31) | 6.5% (K219Q) | None | None | [127] | ||||||
| Gabon | 2000 | 35 (13) | None | None | None | [170] | ||||||
| Ghana | 2001–2002 | 39 (39) | Not studied | Not studied | None | [125] | ||||||
| Ghana | 2003 | 25 (25) | None | None | None | [171] | ||||||
| Kenya | 1999–2000 | 41 (41§) | 7.3§ (M184I) | None§ | 4.9§ (M46L, G73S) | [172] | ||||||
| Kenya | 1995 | 460 (38§) | None§ | 2.6§ (G190A) | 2.6§ (L33F) | [173] | ||||||
| Madagascar | 2005 | 28 (23) | None | None | 4.3 (M46I, I84V, L90M) | [174] | ||||||
| Malawi | 1996–2001 | 21 (21) | None | None | None | [175] | ||||||
| Mali | 2006 | 198 (193) | 1.5% (V75I, K219Q) | 9.0 (V108I, Y181C) | 1.0 (L33F, M46L) | [176,177] | ||||||
| Mozambique | 2008‡ | 75 (75) | None | None | None | [112] | ||||||
| Mozambique | 2003 | 59 (58) | None | None | 1.7 (I50L) | [115] | ||||||
| Mozambique | 1999–2004 | 81 (81) | Not studied | Not studied | None | [178] | ||||||
| Nigeria | 2006‡ | 18 (18) | None | None | None | [114] | ||||||
| Nigeria | 2005 | 50 (43) | 8.6 (M41L) | 2.9 (Y188H) | None | [179] | ||||||
| Rwanda | 2000 | 43 (43) | None | None | 2.3 (L33F) | [180] | ||||||
| Senegal | 1998–2001 | 80 (32§) | None | None | None | [181] | ||||||
| Seychelles | 2005–2006 | 40 (31) | None | None | None | [182] | ||||||
| South Africa | 2003 | 14 (14) | None | 21.0 (K103N, V106M) | Not studied | [72] | ||||||
| South Africa | 2001 | 42 (13) | None | None | None | [69] | ||||||
| South Africa | 2001–2004 | 53 (40) | None | None | 5.0 (M46L, G73S) | [116] | ||||||
| South Africa | 2001–2002 | 72 (61) | None | 3.3 (K103N, G190A) | None | [70] | ||||||
| South Africa | 2000 | 37 (37) | None | None | Not studied | [71] | ||||||
| Swaziland | 2002–2003 | 47 (47) | None | 2.0 (Y181I) | None | [117] | ||||||
| Swaziland | 2006 | 70 (60) | None | None | 3.3 (M46I, I47V) | [183] | ||||||
| Tanzania | 2001 | 36 (20§) | None§ | None§ | None§ | [184] | ||||||
| Tanzania | 2002–2003 | 507 (14§) | 7.1§ (M184I) | 7.1§ (Y188H) | 7.1§ (D30N) | [185] | ||||||
| Tanzania | 2005 | 246 (100§) | 1.0§ (T69D) | 1.0§ (P225H) | 3.0 (L23I, L33F) | [186] | ||||||
| Tanzania | 2005–2006 | 60 (39) | None | None | None | [187] | ||||||
| Uganda | 1993–1994 | 27 (27) | None | None | 3.7 (L33F) | [188] | ||||||
| Uganda | 2007‡ | 279 (254) | 0.8 (M41L) | None | None | [189] | ||||||
| Uganda | 1990 | 187 (187) | None | None | None | [190] | ||||||
| Uganda | 2006–2007 | 46 (46) | None | None | None | [191] | ||||||
| Zambia | 2000 | 28 (28) | None | None | None | [122] | ||||||
| Zimbabwe | 1995 | 12 (12) | Not studied | Not studied | None | [192] | ||||||
| Asia | ||||||||||||
| Azerbaijan | 1999–2002 | 125 (37) | 18.9§ (A62V) | None§ | None§ | [193] | ||||||
| Cambodia | 2003–2004 | 146 (142) | 2.8 (K70R, V75M) | None | 2.8 (L33F, M46I, N88D) | [81] | ||||||
| China | 2005–2006 | 13 (10§) | 10.0§ (M184I) | 20.0§ (Y188L) | 20.0§ (M46I, F53L) | [194] | ||||||
| China | 2000 | 126 (84) | None | None | 1.2 (V82A) | [195] | ||||||
| China | 2003‡ | 66 (52) | None | None | None | [196] | ||||||
| China | 1999–2004 | 91 (38) | 5.3 (M41L, M184I) | None | 5.3 (M46I) | [76] | ||||||
| China | 2003–2004 | 25 (25) | None | None | None | [77] | ||||||
| China | 1999–2001 | 40 (16) | Not reported | Not reported | 6.3 (M46I) | [78] | ||||||
| China | 2005 | 95 (54) | 3.7 (A62V, D67G) | None | 1.9 (M46I) | [79] | ||||||
| Georgia | 1998–2003 | 48 (36) | 2.8 (M184V/I) | None | None | [97] | ||||||
| India | 2003 | 128 (128) | 1.6 (M184V) | None | 0.8 (M46I) | [88] | ||||||
| India | 1999–2001 | 12 (12) | None | None | None | [89] | ||||||
| India | 2004–2005 | 75 (25) | None | None | None | [92] | ||||||
| India | 2007‡ | 48 (48) | None | None | None | [94] | ||||||
| India | 2005‡ | 50 (50) | 2.0 (D67E) | 2.0 (K103N) | 2.0 (M46I) | [95] | ||||||
| India, North | 2008‡ | 52 (52) | None | None | 2.0 (M46I) | [87] | ||||||
| India, South | 2004–2005 | 38 (38) | Not studied | Not studied | None | [90] | ||||||
| India, Western | 2007 | 40 (40) | 7.5 (M41L, D67N, M184V, K219R) | None | 2.5 (V82A) | [91] | ||||||
| Iran | 2009‡ | 13 (13§) | None§ | None§ | None§ | [93] | ||||||
| Israel | 1999–2003 | 176 (147) | 1.4 (M41L, T215Y) | 4.1 (K103N, V106M, V108I, G190A) | 4.8 (M46I, N88D, L90M) | [197] | ||||||
| Japan | 2003–2004 | 575 (97) | None | None | 1.0 (L33F) | [120] | ||||||
| Kazakhstan | 2001–2003 | 85 (85) | 56.5 (A62V) | None | None | [198] | ||||||
| Malaysia | 2003–2004 | 100 (88) | None | 1.0 (Y181C) | 3.4 (L33F) | [83,199] | ||||||
| Moldova | 1997–1998 | 83 (82) | None | None | None | [96] | ||||||
| Singapore | 2002–2003 | 104 (35) | None | None | 2.9 (L33F) | [80] | ||||||
| Soviet Union | 1995–2003 | 119 (119) | Not studied | Not studied | None | [200] | ||||||
| Soviet Union | 1997–2004 | 278 (268) | 8.2 (A62V, V75I, M184V, T215F, K219N/R) | 1.1 (Y188C) | 2.6 (M46I) | [99] | ||||||
| Taiwan | 1999–2006 | 786 (167) | 7.8 total | [86] | ||||||||
| Thailand | 2008‡ | 113 (92) | 6.5 (M41L, T215Y) | None | None | [128] | ||||||
| Thailand | 2006–2007 | 11 (7) | None | None | None | [84] | ||||||
| Thailand | 2000–2001 | 21 (20) | None | None | None | [85] | ||||||
| Thailand | 1999–2002 | 39 (38§) | 13.2§ (V75M, M184I) | None§ | 5.3§ (D30N, M46I, G73S) | [74] | ||||||
| Thailand | 1998–2001 | 168 (25§) | 16.0§ (M184I) | None§ | 24.0§ (D30N, M46I, G73S, I84V) | [75] | ||||||
| Ukraine | 2001–2002 | 163 (114) | 4.4§ (A62V, M184I) | 0.9§ (Y181I) | 0.9§ (M46I) | [201] | ||||||
| Uzbekistan | 2002–2003 | 142 (140) | 65.6 (A62V) | None | None | [202] | ||||||
| Vietnam | 2007 | 301 (291) | 0.7 (M184I, K219E) | 1.8 (K103N, G190E) | 0.3 (M46I) | [82] | ||||||
| Vietnam | 1998–2000 | 25 (23) | None | None | None | [203] | ||||||
| Vietnam | 2001–2002 | 200 (199) | 6.0 (M41L, M184I, K219Q/N) | None | 3.0 (D30N, L33F, M46I, L90M) | [204] | ||||||
| Yemen | 2000–2002 | 19 (10) | None | None | None | [205] | ||||||
| Europe | ||||||||||||
| Albania | 1994–2003 | 72 (43) | 2.3 (M41L, D67N, T69D, T215D) | None | None | [206] | ||||||
| Belgium | 1983–2001 | 281 (41§) | 2.4§ (L74V) | None§ | None§ | [67] | ||||||
| Belgium | 2003–2006 | 285 (117) | 2.6 (M184V, K219E) | 2.6 (K103N, G190A, Y181C) | 1.7 (L90M) | [111] | ||||||
| Cyprus | 2003–2006 | 37 (24) | 4.2 (M184V) | None | None | [110] | ||||||
| Denmark | 2000 | 104 (40) | None | None | None | [109] | ||||||
| Europe | 1996–2002 | 2208 (673) | 4.8 Total | [12] | ||||||||
| France | 1999–2001 | 72 (72) | 4.2 (D67N, K70R, M184I) | 1.4 (K103N) | 1.4 (L33F) | [118] | ||||||
| Germany | 2001–2003 | 346 (76) | 1.4 total (nonsubtype B-specific mutations were not reported) | [59] | ||||||||
| Greece | 2002–2003 | 101 (53) | 1.9 (A62V) | 3.8 (V108I, Y181C) | None | [207] | ||||||
| Italy | 2004–06 | 111 (13) | None | None | None | [121] | ||||||
| Portugal | 2003 | 180 (105) | 5.7% (M41L, M184V, K219Q, T215C) | 2.9 (K103N, V108I, G190A) | 1.0 (L90M) | [208] | ||||||
| Portugal | 1998–2000 | 52 (52) | Not studied | Not studied | 11.5§ (V32I, D30N, I47V, F53L, N88S) | [209] | ||||||
| Romania | 2007 | 29 (29) | None | None | None | [107] | ||||||
| Slovenia | 2000–2004 | 77 (14) | None | None | None | [210] | ||||||
| Spain | 1986–2000 | 141 (71) | Not studied | Not studied | None | [211] | ||||||
| Spain | 2000–2002 | 85 (19) | None | None | 5.3 (M46I) | [212] | ||||||
| Sweden | 1998–2001 | 100 (45) | None | None | 2.2 (M46I) | [213] | ||||||
| Switzerland | 1996–2005 | 822 (241) | 3.7 total (nonsubtype B-specific mutations were not reported) | [14] | ||||||||
| UK | 2004–2006 | 239 (105) | None | 1.0 (K103N) | 1.0 (M46L) | [108] | ||||||
| UK | 1996–2003 | 2357 (424) | 13.4 total | [13] | ||||||||
| Multiple | 1997–2002 | 58 (58) | Not studied | Not studied | None | [214] | ||||||
| Multiple | 1986–1998 | 301 (187) | Not studied | Not studied | 2.1 (D30N, M46L, V82F, I85V) | [54] | ||||||
| North America | ||||||||||||
| USA (Boston) | 1999 | 115 (9) | None | None | None | [65] | ||||||
| USA (NYC) | 2000–2004 | 151 (9) | 11.1 (K219Q) | 11.1 (K103N) | None | [66] | ||||||
| USA | 1997–2000 | 520 (12§) | 8.3§ (M184I) | None§ | 15.4§ (D30N, M46I, G73S) | [215] | ||||||
| Central America | ||||||||||||
| Cuba | 2003 | 425 (141) | 1.4 (K70R, M184V, T215D) | None | None | [103] | ||||||
| South America | ||||||||||||
| Argentina | 2003–2005 | 323 (156) | 1.9 (M41L, M184V, L210W) | 1.9 (M46L, V82A) | 2.6 (V108I, K103N, Y181C) | [104] | ||||||
| Argentina | 2004–2005 | 52 (31) | None | 3.2 (K103N) | None | [105] | ||||||
| Brazil | 1998–2002 | 648 (64) | 6.3 (M41L, A62V, V75A, M184V, T215F) | None | 3.1 (M46L, V82A) | [106] | ||||||
| Brazil | 2005‡ | 108 (73) | None | None | None | [100] | ||||||
| Brazil | 2000–2004 | 74 (12) | None | None | None | [101] | ||||||
| Brazil | 2002 | 77 (6) | None | None | None | [102] | ||||||
Resistance (%) and specific mutations are based on data reported in the given study that appear on the surveillance drug-resistance mutation list or that are considered a major mutation by the International AIDS Society list. The data may or may not agree with the reported study depending on which list of mutations the authors referred to.
The exact year of the sample population is unknown. Date refers to year of publication and/or the year sequences were deposited in Genbank.
Publications in which resistance data are unclear or not evaluated. Resistance data are gathered from sequences deposited in the Stanford Database [303]. CAR: Central African Republic; DRC: Democratic Republic of Congo; NNRTI: Non-nucleoside reverse-transcriptase inhibitor; NRTI: Nucleoside reverse-transcriptase inhibitor; NYC: New York City; PI: Protease inhibitor; TDR: Transmitted drug resistance.
The prevalence of TDR also varies in Asia [74,75]. The rate of reported TDR mutations in nonsubtype B viruses in China is low, generally less than 6% [76–79], and similar to Singapore [80], Cambodia [81], Vietnam [82] and Malaysia [83]. In Thailand, reported resistance ranged from 0 [84,85] up to 24% [74,75]. This high rate was due to RT mutation M184I and protease mutations D30N, M46I, G73S and I84V, reported yet again from a limited sample size (n = 25). Although subtype B is predominant in Taiwan, the rates of resistance among non-B subtypes was reported as 8% in samples collected from 1999 to 2006 [86]. In India, where subtype C is the dominant subtype, reported resistance is less than 5% [87–95]. In Russia and the former Soviet Union, subtype A and CRF01_AE predominate. Reported TDR ranges from 0% in Moldova [96] and 3% in the Republic of Georgia [97] to 7% in Latvia [98] and 8% in a multinational study of this region [99]. The rate of 8% included A62V, which is not considered a marker of TDR according to SDRM.
Although much of central and South America is populated by HIV-1 subtype B, reports of non-B subtypes are increasing and several studies assessed the prevalence of TDR in nonsubtype B infections. Reports range from 0 to 1% in Brazil [100–102] and Cuba [103], to 3% in Argentina [104,105] and as high as 9% in Brazil [106].
In Europe, nonsubtype B viruses make up a substantial proportion of HIV-1 infections (up to 100% subtype F1 in parts of Romania [107]) and is increasingly prevalent in other areas of the continent [12–14,108–111]. From 1996 to 2002 in several countries in Europe, the overall prevalence of TDR among 673 nonsubtype B individuals was 5% [12]. In the USA, although the reported population of nonsubtype B is much smaller than Europe, [65,66], TDR has ranged from 0% in 1999 in Boston (MA, USA) [65] to more than 20% in 2000–2004 in New York City (NY, USA) [66]. Populations from these regions, with long-term antiretroviral access, have higher rates of TDR. Furthermore, rates of resistance to NRTIs and NNRTIs are higher than those for PIs, coinciding with the greater use of these drugs and their universal earlier introduction.
Mutations associated with TDR in nonsubtype B HIV-1
Transmitted drug-resistance mutations were reported at 37 different positions in the RT and protease genes in non-B HIV-1 subtypes and recombinants across the three classes of antiretroviral drugs (12 NRTI, ten NNRTI and 15 PI positions associated with resistance; Table 3). The most commonly reported NRTI TDR mutations were M184V/I. Additional common mutations include M41L, K219E/N/Q/R and T215F/Y/C/D. M184V, observed across numerous populations and subtypes, confers high-level resistance to lamivudine and emtricitabine. The additional reported mutations are termed thymidine analog mutations and can confer resistance to all NRTIs. The most commonly reported NNRTI TDR mutations were K103N, Y181C/I and Y188C/H/L, all conferring resistance across the NNRTI class. RT mutations A62V (commonly reported in subtype A from central Asia) and V108I (commonly reported from Africa) are not on the SDRM list, but are on the IAS-USA list.
Table 3. Reverse transcriptase and protease drug-resistance mutations according to subtype from studies on transmitted resistance in HIV-1 nonsubtype B-infected treatment-naive patients.
| Subtype or recombinant form* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A n = 1755 |
C n = 2423 |
D n = 447 |
F n = 145 |
G n = 420 |
HIJK n = 65 |
01_AE n = 1238 |
02_AG n = 1243 |
Other‡ n = 1810 |
Unk§ n = 435 |
Total (%) n = 9984 |
|
| Nucleoside reverse-transcriptase inhibitors mutations | |||||||||||
| M41 | L3 | L3 | L1 | L1 | L4 | L1 | L6 | 19 (3.6) | |||
| A62¶ | V179 | V2 | V2 | V3 | V1 | 187 (35.3) | |||||
| D67 | N1 | E1N1 | N1 | E1G1 | 6 (1.1) | ||||||
| T69 | D1 | D1 | 2 (0.4) | ||||||||
| K70 | ER2 | R1 | R2 | 5 (0.9) | |||||||
| L74 | V1 | I1 | 2 (0.4) | ||||||||
| V75 | I1 | A1 | I1 | M4 | I1 | I1 | 9 (1.7) | ||||
| F77 | L2 | 2 (0.4) | |||||||||
| M184 | I3V6 | I1V3 | V1 | I14 | I1 | I2V5 | V1 | 37 (7.0) | |||
| L210 | W2 | W1 | W4 | 7 (1.3) | |||||||
| T215 | F1 | FY2 | D1 | F1 | Y3 | YF2 | C1D1 | 12 (2.3) | |||
| K219 | Q1NR2 | E1Q2 | E1QN7 | Q2 | Q2 | 18 (3.4) | |||||
| Non-nucleoside reverse-transcriptase inhibitors mutations | |||||||||||
| L100 | I1 | I1 | 2 (0.4) | ||||||||
| K101 | E1 | E1 | 2 (0.4) | ||||||||
| K103 | N3 | N10 | N3 | N3 | N5 | N2 | 26 (4.9) | ||||
| V106 | M2 | A1 | 3 (0.6) | ||||||||
| V108¶ | I3 | I1 | I1 | I1 | I3 | I5 | I4 | 18 (3.4) | |||
| Y181 | C2I1 | I1 | C1 | C1 | C7 | 13 (2.5) | |||||
| Y188 | C1H1 | H1 | H1 | L2 | C1 | C5CH3 | 15 (2.8) | ||||
| G190 | A5E1 | A1 | E1 | E1 | 9 (1.7) | ||||||
| P225 | H1 | 1 (0.2) | |||||||||
| M230 | L1 | 1 (0.2) | |||||||||
| Protease inhibitors mutations | |||||||||||
| L23 | I1 | 1 (0.2) | |||||||||
| L24 | I1 | 1 (0.2) | |||||||||
| D30 | N1 | N4 | N1 | N5 | 11 (2.1) | ||||||
| V32 | I1 | 1 (0.2) | |||||||||
| L33¶ | F3 | F1 | F2 | F6 | F6 | F1 | F3 | 22 (4.2) | |||
| M46 | I8L4 | I8L1 | I2L2 | I3 | I13 | I1L1 | I3L6 | I/L4 | 56 (10.6) | ||
| I47 | V1 | V2 | 3 (0.6) | ||||||||
| I50 | L1 | 1 (0.2) | |||||||||
| F53 | L1 | L1 | L1 | Y1 | 4 (0.8) | ||||||
| G73 | S1 | S1 | S4 | S2 | 8 (1.5) | ||||||
| V82 | A1F1 | A2 | A1 | A1 | 6 (1.1) | ||||||
| I84 | V1 | V1 | 2 (0.4) | ||||||||
| I85 | V1 | 1 (0.2) | |||||||||
| N88 | D1S1 | D1 | D3S1 | 7 (1.3) | |||||||
| L90 | M1 | M1 | M1 | M3 | M3 | 9 (1.7) | |||||
| Total (%) | 227 (12.9) | 62 (2.6) | 13 (2.9) | 10 (6.9) | 16 (3.8) | 1 (1.5) | 79 (6.4) | 37 (3.0) | 68 (3.8) | 16 (3.7) | 529 (5.3) |
Left column indicates wild-type amino acid (for consensus subtype B sequence) and reverse transcriptase or protease position. Other columns indicate mutation and number of occurrences (subscript) present per non-B subtype.
Includes recombinant and mosaic species.
Includes mutations in non-B subtypes that were not differentiated by subtype.
Mutations are not on the surveillance drug-resistance mutation list, but are considered major resistance mutations according to the International AIDS Society–USA.
A: Alanine; C: Cysteine; D: Aspartic acid; E: Glutamic acid; F: Phenylalanine; G: Glycine; H: Histidine; I: Isoleucine; K: Lysine; L: Leucine; M: Methionine; N: Asparagine; P: Proline; Q: Glutamine; R: Arginine; S: Serine; T: Threonine; Unk: Unknown; V: Valine; W: Tryptophan; Y: Tyrosine.
The most commonly reported PI TDR mutations were M46I/L. Mutations at this position were not included in the 2007 SDRM list [51] but are part of the 2009 SDRM list (Table 1) [45]. Other TDR PI mutations were reported much less frequently, as expected by less drug exposure owing to the inclusion of mostly RT inhibitors in first-line regimens in resource-limited settings. L33F, reported from Africa and Asia, appears on the IAS-USA list and not on the SDRM list, similar to RT mutations A62V and V108I mentioned earlier. Although major PI resistance mutations were less common than mutations in RT, prevalence of accessory protease mutations was abundant, as discussed previously [54,80,81,89,95,105,107,110,112–129]. Compared with more accepted TDR mutations, these polymorphisms appear to have minimal effect on treatment outcomes [8,128–134]; however, some HAART regimens may be less efficacious [118,135]. Although the genetic barrier (or the number of genetic changes necessary to create drug-resistance mutations) appears to be similar for different subtypes despite baseline genotypic difference [136], the significance of these pretherapy polymorphisms in the evolution and transmission of drug resistance is still unclear [39].
As demonstrated in Table 3, the number of TDR mutations reported in the studies reviewed here is small. Overall, in 9984 non-B sequences, there were 529 (5.3%) drug-resistance mutations. Upon exclusion of mutations that appear on the IAS-USA list as major mutations but not on the SDRM list (RT A62V and V108I; protease L33F), that number declines to 302 mutations (3%). When specific subtypes are examined, other than subtype F and CRF01_AE, all TDR rates are below 5%. Whether the 6.9% TDR in subtype F and 6.4% TDR in CRF01_AE (5.7% without non-SDRM mutations) are actual differences will need to be examined in larger data sets. Regarding specific TDR mutations, despite the small numbers, some observations can be made, such as the lack of M41L in 1755 subtype A sequences; the lack of K103N in subtypes D, F and G sequences; and isoleucine as the only mutation at RT position 184 in CRF01_AE. Whether these observations are significant remains to be determined. Submission of sequence data to electronic databases and clear presentations of study methodologies will allow further careful examinations of TDR in multiple HIV-1 subtypes.
Conclusion
Infections with nonsubtype B HIV-1 predominate globally and are on the rise in geographic areas where subtype B prevails, such as North America and Europe. The epidemiology of HIV drug resistance in resource-rich settings is complex and with increased universal access to antiretroviral therapy, we are only now beginning to examine it in resource-limited settings. TDR is important to determine as it results in longer time to viral suppression and shorter time to virological failure. High rates of TDR have been reproted from regions in which antiretroviral therapy has been available for a long time and, therefore, it is likely that it will increase in areas where treatment access is being scaled-up.
Attempts to standardize global TDR surveillance strategies are ongoing. This should include quantifying TDR and identifying mutations that accurately represent transmission of drug resistance and not merely part of HIV diversity. Methodologies to determine TDR in nonsubtype B HIV-1 vary widely across different settings, and should be carefully interpreted. Additional limitations in the ability to aggregate and interpret reported data on TDR in non-B subtypes include resistance transmission not being the main study focus, variable subtyping methods and paucity of data.
Executive summary.
HIV diversity
Nonsubtype B HIV-1 variants account for the majority of infections worldwide and vary according to group, subtype and recombinant form.
These variants are increasing in areas where subtype B infection currently predominates.
Transmitted drug resistance
Transmitted drug resistance (TDR) is defined as mutations that confer resistance to one or more antiretroviral drugs found in individuals with no previous drug exposure.
TDR is attributed to direct transmission of resistant viral strains from treated to untreated individuals.
TDR to protease inhibitors, nucleoside reverse-transcriptase inhibitors and non-nucleoside reverse-transcriptase inhibitors is being reported globally, and is higher in areas with longer exposure to antiretroviral treatment.
TDR in nonsubtype B HIV-1
Definition, detection, interpretation and reporting of TDR are variable and should be carefully examined in the context of study design and HIV diversity.
Different lists of drug-resistance mutations that lack consensus lead to TDR reporting and resistance interpretation discordances.
Standardization attempts and global surveillance efforts are ongoing.
Mutations that appear as natural polymorphisms, and that are common in non-B HIV-1 subtypes regardless of drug resistance transmission, are often reported as markers of TDR and lead to its overestimation.
Additional data and more explicit descriptions of study methodologies will allow further examination of TDR in HIV-1 subtypes.
Future perspective
An interesting and challenging question is how long after the introduction of antiretroviral therapy should TDR be expected to develop and at what pace. Mathematical models were shown to be fairly accurate in predicting rates of TDR in certain populations in North America [137,138], and estimations regarding other global settings are ongoing [139,140]. Although prolonged access to therapy and the use of less potent regimens appear to be linked to high TDR prevalence, this observation is not always consistent. Important effects seem to include the proportion of antiretroviral treatment in the population examined, the development of acquired resistance in the population, adherence to therapy and viral fitness. How these will vary in different resource-limited settings with diverse HIV-1 subtypes remains to be determined.
Acknowledgments
Rami Kantor is funded by an NIH RO1 grant AI66922.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
 of interest
of interest
 of considerable interest
of considerable interest
- 1.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet. 2003;362:2002–2011. doi: 10.1016/S0140-6736(03)15022-2. [DOI] [PubMed] [Google Scholar]
- 3.Kozal MJ. Drug-resistant human immunodefiency virus. Clin Microbiol Infect. 2009;15(Suppl 1):69–73. doi: 10.1111/j.1469-0691.2008.02687.x. [DOI] [PubMed] [Google Scholar]
- 4.Daar ES. Emerging resistance profiles of newly approved antiretroviral drugs. Top HIV Med. 2008;16:110–116. [PubMed] [Google Scholar]
- 5.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
-
6.Wainberg MA. HIV-1 subtype distribution and the problem of drug resistance. AIDS. 2004;18(Suppl 3):S63–S68. doi: 10.1097/00002030-200406003-00012. [DOI] [PubMed] [Google Scholar];
 Focuses on the susceptibility of different HIV-1 subtypes to antiretroviral therapy and the development of drug resistance.
Focuses on the susceptibility of different HIV-1 subtypes to antiretroviral therapy and the development of drug resistance.
- 7.Kantor R, Katzenstein D. Drug resistance in nonsubtype B HIV-1. J Clin Virol. 2004;29:152–159. doi: 10.1016/S1386-6532(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 8.Deroo S, Robert I, Fontaine E, et al. HIV-1 subtypes in Luxembourg, 1983–2000. AIDS. 2002;16:2461–2467. doi: 10.1097/00002030-200212060-00012. [DOI] [PubMed] [Google Scholar]
-
9.Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society–USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar];
 Current guidelines for drug-resistance testing followed by most institutions in the developed world.
Current guidelines for drug-resistance testing followed by most institutions in the developed world.
- 10.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 11.Wainberg MA, Friedland G. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA. 1998;279:1977–1983. doi: 10.1001/jama.279.24.1977. [DOI] [PubMed] [Google Scholar]
-
12.Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192:958–966. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar];
 Large study comparing the prevalence of transmitted drug resistance (TDR) in subtype B and nonsubtype B HIV-1 in Europe.
Large study comparing the prevalence of transmitted drug resistance (TDR) in subtype B and nonsubtype B HIV-1 in Europe.
- 13.Cane P, Chrystie I, Dunn D, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: multicentre observational study. BMJ. 2005;331:1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yerly S, von Wyl V, Ledergerber B, et al. Transmission of HIV-1 drug resistance in Switzerland: a 10-year molecular epidemiology survey. AIDS. 2007;21:2223–2229. doi: 10.1097/QAD.0b013e3282f0b685. [DOI] [PubMed] [Google Scholar]
- 15.Katabira ET, Oelrichs RB. Scaling up antiretroviral treatment in resource-limited settings: successes and challenges. AIDS. 2007;21(Suppl 4):S5–S10. doi: 10.1097/01.aids.0000279701.93932.ef. [DOI] [PubMed] [Google Scholar]
- 16.Gilks CF, Crowley S, Ekpini R, et al. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006;368:505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 17.Galarraga O, O'Brien ME, Gutierrez JP, et al. Forecast of demand for antiretroviral drugs in low and middle-income countries: 2007–2008. AIDS. 2007;21(Suppl 4):S97–S103. doi: 10.1097/01.aids.0000279712.32051.29. [DOI] [PubMed] [Google Scholar]
- 18.Boyd MA, Cooper DA. Second-line combination antiretroviral therapy in resource-limited settings: facing the challenges through clinical research. AIDS. 2007;21(Suppl 4):S55–S63. doi: 10.1097/01.aids.0000279707.01557.b2. [DOI] [PubMed] [Google Scholar]
- 19.Renaud-Thery F, Nguimfack BD, Vitoria M, et al. Use of antiretroviral therapy in resource-limited countries in 2006: distribution and uptake of first- and second-line regimens. AIDS. 2007;21(Suppl 4):S89–S95. doi: 10.1097/01.aids.0000279711.54922.f0. [DOI] [PubMed] [Google Scholar]
- 20.Hammer SM, Eron JJ, Jr, Reiss P, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society–USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 21.Vandamme AM, Sonnerborg A, Ait-Khaled M, et al. Updated European recommendations for the clinical use of HIV drug resistance testing. Antivir Ther. 2004;9:829–848. [PubMed] [Google Scholar]
- 22.Gazzard BG. British HIV Association Guidelines for the treatment of HIV-1-infected adults with antiretroviral therapy 2008. HIV Med. 2008;9:563–608. doi: 10.1111/j.1468-1293.2008.00636.x. [DOI] [PubMed] [Google Scholar]
-
23.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. [PubMed] [Google Scholar];
 Excellent overview of the international approach to monitoring TDR in resource-poor settings as presented by the WHO.
Excellent overview of the international approach to monitoring TDR in resource-poor settings as presented by the WHO.
- 24.Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 25.Booth CL, Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. J Antimicrob Chemother. 2007;59:1047–1056. doi: 10.1093/jac/dkm082. [DOI] [PubMed] [Google Scholar]
- 26.Shet A, Berry L, Mohri H, et al. Tracking the prevalence of transmitted antiretroviral drug-resistant HIV-1: a decade of experience. J Acquir Immune Defic Syndr. 2006;41:439–446. doi: 10.1097/01.qai.0000219290.49152.6a. [DOI] [PubMed] [Google Scholar]
- 27.Smith D, Moini N, Pesano R, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44:456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 28.Ghosn J, Pellegrin I, Goujard C, et al. HIV-1 resistant strains acquired at the time of primary infection massively fuel the cellular reservoir and persist for lengthy periods of time. AIDS. 2006;20:159–170. doi: 10.1097/01.aids.0000199820.47703.a0. [DOI] [PubMed] [Google Scholar]
- 29.Bon I, Gibellini D, Borderi M, et al. Genotypic resistance in plasma and peripheral blood lymphocytes in a group of naive HIV-1 patients. J Clin Virol. 2007;38:313–320. doi: 10.1016/j.jcv.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Metzner KJ, Rauch P, Walter H, et al. Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS. 2005;19:1819–1825. doi: 10.1097/01.aids.0000189878.97480.ed. [DOI] [PubMed] [Google Scholar]
- 31.Paolucci S, Baldanti F, Campanini G, et al. Analysis of HIV drug-resistant quasispecies in plasma, peripheral blood mononuclear cells and viral isolates from treatment-naive and HAART patients. J Med Virol. 2001;65:207–217. doi: 10.1002/jmv.2022. [DOI] [PubMed] [Google Scholar]
- 32.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 33.Charpentier C, Dwyer DE, Mammano F, Lecossier D, Clavel F, Hance AJ. Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors. J Virol. 2004;78:4234–4247. doi: 10.1128/JVI.78.8.4234-4247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peuchant O, Thiebaut R, Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008;22:1417–1423. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- 35.Johnson JA, Li JF, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 36.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roquebert B, Malet I, Wirden M, et al. Role of HIV-1 minority populations on resistance mutational pattern evolution and susceptibility to protease inhibitors. AIDS. 2006;20:287–289. doi: 10.1097/01.aids.0000202650.03279.69. [DOI] [PubMed] [Google Scholar]
- 38.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19:594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- 40.Kantor R, Katzenstein D. Polymorphism in HIV-1 nonsubtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 2003;5:25–35. [PubMed] [Google Scholar]
- 41.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 42.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 43.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 44.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- 45.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravela J, Betts BJ, Brun-Vezinet F, et al. HIV-1 protease and reverse transcriptase mutation patterns responsible for discordances between genotypic drug resistance interpretation algorithms. J Acquir Immune Defic Syndr. 2003;33:8–14. doi: 10.1097/00126334-200305010-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt B, Walter H, Zeitler N, Korn K. Genotypic drug resistance interpretation systems – the cutting edge of antiretroviral therapy. AIDS Rev. 2002;4:148–156. [PubMed] [Google Scholar]
- 48.Snoeck J, Kantor R, Shafer RW, et al. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother. 2006;50:694–701. doi: 10.1128/AAC.50.2.694-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturmer M, Doerr HW, Preiser W. Variety of interpretation systems for human immunodeficiency virus type 1 genotyping: confirmatory information or additional confusion? Curr Drug Targets Infect Disord. 2003;3:373–382. doi: 10.2174/1568005033481006. [DOI] [PubMed] [Google Scholar]
-
50.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE 4. 2009:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar];
 Current surveillance drug-resistance mutation list for the definition of TDR mutations.
Current surveillance drug-resistance mutation list for the definition of TDR mutations.
- 51.Shafer RW, Rhee SY, Pillay D, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clemente JC, Hemrajani R, Blum LE, Goodenow MM, Dunn BM. Secondary mutations M36I and A71V in the human immunodeficiency virus type 1 protease can provide an advantage for the emergence of the primary mutation D30N. Biochemistry. 2003;42:15029–15035. doi: 10.1021/bi035701y. [DOI] [PubMed] [Google Scholar]
- 53.Cornelissen M, van den Burg R, Zorgdrager F, Lukashov V, Goudsmit J. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pieniazek D, Rayfield M, Hu DJ, et al. Protease sequences from HIV-1 group M subtypes A–H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working Group AIDS. 2000;14:1489–1495. doi: 10.1097/00002030-200007280-00004. [DOI] [PubMed] [Google Scholar]
-
55.Green H, Tilston P, Fearnhill E, Pillay D, Dunn DT. The impact of different definitions on the estimated rate of transmitted HIV drug resistance in the United Kingdom. J Acquir Immune Defic Syndr. 2008;49:196–204. doi: 10.1097/QAI.0b013e318185725f. [DOI] [PubMed] [Google Scholar];
 TDR comparison according to different mutation lists in a large UK population.
TDR comparison according to different mutation lists in a large UK population.
- 56.Eshleman SH, Husnik M, Hudelson S, et al. Antiretroviral drug resistance, HIV-1 tropism, and HIV-1 subtype among men who have sex with men with recent HIV-1 infection. AIDS. 2007;21:1165–1174. doi: 10.1097/QAD.0b013e32810fd72e. [DOI] [PubMed] [Google Scholar]
- 57.Viani RM, Peralta L, Aldrovandi G, et al. Prevalence of primary HIV-1 drug resistance among recently infected adolescents: a multicenter adolescent medicine trials network for HIV/AIDS interventions study. J Infect Dis. 2006;194:1505–1509. doi: 10.1086/508749. [DOI] [PubMed] [Google Scholar]
- 58.Corvasce S, Violin M, Romano L, et al. Evidence of differential selection of HIV-1 variants carrying drug-resistant mutations in seroconverters. Antivir Ther. 2006;11:329–334. [PubMed] [Google Scholar]
- 59.Oette M, Kaiser R, Daumer M, et al. Primary HIV drug resistance and efficacy of first-line antiretroviral therapy guided by resistance testing. J Acquir Immune Defic Syndr. 2006;41:573–581. doi: 10.1097/01.qai.0000214805.52723.c1. [DOI] [PubMed] [Google Scholar]
- 60.Chaix ML, Descamps D, Wirden M, et al. Stable frequency of HIV-1 transmitted drug resistance in patients at the time of primary infection over 1996–2006 in France. AIDS. 2009;23:717–724. doi: 10.1097/QAD.0b013e328326ca77. [DOI] [PubMed] [Google Scholar]
- 61.Alvarez M, Garcia F, Martinez NM, et al. Introduction of HIV type 1 non-B subtypes into Eastern Andalusia through immigration. J Med Virol. 2003;70:10–13. doi: 10.1002/jmv.10368. [DOI] [PubMed] [Google Scholar]
- 62.Balotta C, Facchi G, Violin M, et al. Increasing prevalence of non-clade B HIV-1 strains in heterosexual men and women, as monitored by analysis of reverse transcriptase and protease sequences. J Acquir Immune Defic Syndr. 2001;27:499–505. doi: 10.1097/00126334-200108150-00012. [DOI] [PubMed] [Google Scholar]
- 63.Brennan CA, Stramer SL, Holzmayer V, et al. Identification of human immunodeficiency virus type 1 non-B subtypes and antiretroviral drug-resistant strains in United States blood donors. Transfusion. 2009;49:125–133. doi: 10.1111/j.1537-2995.2008.01935.x. [DOI] [PubMed] [Google Scholar]
- 64.Frange P, Galimand J, Vidal N, et al. New and old complex recombinant HIV-1 strains among patients with primary infection in 1996–2006 in France: the French ANRS CO06 primo cohort study. Retrovirology. 2008;5:69. doi: 10.1186/1742-4690-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanna GJ, Balaguera HU, Freedberg KA, et al. Drug-selected resistance mutations and non-B subtypes in antiretroviral-naive adults with established human immunodeficiency virus infection. J Infect Dis. 2003;188:986–991. doi: 10.1086/378280. [DOI] [PubMed] [Google Scholar]
- 66.Parker MM, Gordon D, Reilly A, et al. Prevalence of drug-resistant and nonsubtype B HIV strains in antiretroviral-naive, HIV-infected individuals in New York State. AIDS Patient Care STDS. 2007;21:644–652. doi: 10.1089/apc.2006.0172. [DOI] [PubMed] [Google Scholar]
- 67.Snoeck J, Van Laethem K, Hermans P, et al. Rising prevalence of HIV-1 non-B subtypes in Belgium: 1983–2001. J Acquir Immune Defic Syndr. 2004;35:279–285. doi: 10.1097/00126334-200403010-00009. [DOI] [PubMed] [Google Scholar]
- 68.Tramuto F, Vitale F, Bonura F, Romano N. Detection of HIV type 1 non-B subtypes in Sicily, Italy. AIDS Res Hum Retroviruses. 2004;20:251–254. doi: 10.1089/088922204773004987. [DOI] [PubMed] [Google Scholar]
- 69.Bessong PO, Larry Obi C, Cilliers T, et al. Characterization of human immunodeficiency virus type 1 from a previously unexplored region of South Africa with a high HIV prevalence. AIDS Res Hum Retroviruses. 2005;21:103–109. doi: 10.1089/aid.2005.21.103. [DOI] [PubMed] [Google Scholar]
- 70.Gordon M, De Oliveira T, Bishop K, et al. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies. J Virol. 2003;77:2587–2599. doi: 10.1128/JVI.77.4.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pillay C, Bredell H, McIntyre J, Gray G, Morris L. HIV-1 subtype C reverse transcriptase sequences from drug-naive pregnant women in South Africa. AIDS Res Hum Retroviruses. 2002;18:605–610. doi: 10.1089/088922202753747950. [DOI] [PubMed] [Google Scholar]
- 72.Barth RE, Wensing AM, Tempelman HA, Moraba R, Schuurman R, Hoepelman AI. Rapid accumulation of nonnucleoside reverse transcriptase inhibitor-associated resistance: evidence of transmitted resistance in rural South Africa. AIDS. 2008;22:2210–2212. doi: 10.1097/QAD.0b013e328313bf87. [DOI] [PubMed] [Google Scholar]
- 73.Koch N, Ndihokubwayo JB, Yahi N, Tourres C, Fantini J, Tamalet C. Genetic analysis of hiv type 1 strains in Bujumbura (Burundi): predominance of subtype c variant. AIDS Res Hum Retroviruses. 2001;17:269–273. doi: 10.1089/088922201750063205. [DOI] [PubMed] [Google Scholar]
- 74.Tovanabutra S, Beyrer C, Sakkhachornphop S, et al. The changing molecular epidemiology of HIV type 1 among northern Thai drug users, 1999 to 2002. AIDS Res Hum Retroviruses. 2004;20:465–475. doi: 10.1089/088922204323087705. [DOI] [PubMed] [Google Scholar]
- 75.Watanaveeradej V, Benenson MW, Souza MD, et al. Molecular epidemiology of HIV type 1 in preparation for a Phase III prime-boost vaccine trial in Thailand and a new approach to HIV type 1 genotyping. AIDS Res Hum Retroviruses. 2006;22:801–807. doi: 10.1089/aid.2006.22.801. [DOI] [PubMed] [Google Scholar]
- 76.Han X, Zhang M, Dai D, et al. Genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV/AIDS patients living in Liaoning Province, China: baseline prevalence and subtype-specific difference. AIDS Res Hum Retroviruses. 2007;23:357–364. doi: 10.1089/aid.2006.0094. [DOI] [PubMed] [Google Scholar]
- 77.Liao L, Xing H, Li X, et al. Genotypic analysis of the protease and reverse transcriptase of HIV type 1 isolates from recently infected injecting drug users in western China. AIDS Res Hum Retroviruses. 2007;23:1062–1065. doi: 10.1089/aid.2007.0050. [DOI] [PubMed] [Google Scholar]
- 78.Zhong P, Kang L, Pan Q, et al. Identification and distribution of HIV type 1 genetic diversity and protease inhibitor resistance-associated mutations in Shanghai, P. R. China. J Acquir Immune Defic Syndr. 2003;34:91–101. doi: 10.1097/00126334-200309010-00014. [DOI] [PubMed] [Google Scholar]
- 79.Zhong P, Pan Q, Ning Z, et al. Genetic diversity and drug resistance of human immunodeficiency virus type 1 (HIV-1) strains circulating in Shanghai. AIDS Res Hum Retroviruses. 2007;23:847–856. doi: 10.1089/aid.2006.0196. [DOI] [PubMed] [Google Scholar]
- 80.Hsu LY, Subramaniam R, Bacheler L, Paton NI. Characterization of mutations in CRF01_AE virus isolates from antiretroviral treatment-naive and -experienced patients in Singapore. J Acquir Immune Defic Syndr. 2005;38(1):5–13. doi: 10.1097/00126334-200501010-00002. [DOI] [PubMed] [Google Scholar]
- 81.Ly N, Recordon-Pinson P, Phoung V, et al. Characterization of mutations in HIV type 1 isolates from 144 Cambodian recently infected patients and pregnant women naive to antiretroviral drugs. AIDS Res Hum Retroviruses. 2005;21:971–976. doi: 10.1089/aid.2005.21.971. [DOI] [PubMed] [Google Scholar]
- 82.Ishizaki A, Cuong NH, Thuc PV, et al. Profile of HIV type 1 infection and genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV type 1-infected individuals in Hai Phong, Vietnam. AIDS Res Hum Retroviruses. 2009;25:175–182. doi: 10.1089/aid.2008.0193. [DOI] [PubMed] [Google Scholar]
- 83.Tee KK, Kamarulzaman A, Ng KP. Short communication: low prevalence of genotypic drug resistance mutations among antiretroviral-naive HIV type 1 patients in Malaysia. AIDS Res Hum Retroviruses. 2006;22:121–124. doi: 10.1089/aid.2006.22.121. [DOI] [PubMed] [Google Scholar]
- 84.Ananworanich J, Phanuphak N, de Souza M, et al. Incidence and characterization of acute HIV-1 infection in a high-risk Thai population. J Acquir Immune Defic Syndr. 2008;49:151–155. doi: 10.1097/QAI.0b013e318183a96d. [DOI] [PubMed] [Google Scholar]
- 85.Auswinporn S, Jenwitheesuk E, Panburana P, Sirinavin S, Vibhagool A, Chantratita W. Prevalence of HIV-1 drug resistance in antiretroviral-naive pregnant Thai women. Southeast Asian J Trop Med Public Health. 2002;33:818–821. [PubMed] [Google Scholar]
- 86.Chang SY, Chen MY, Lee CN, et al. Trends of antiretroviral drug resistance in treatment-naive patients with human immunodeficiency virus type 1 infection in Taiwan. J Antimicrob Chemother. 2008;61:689–693. doi: 10.1093/jac/dkn002. [DOI] [PubMed] [Google Scholar]
- 87.Arora SK, Gupta S, Toor JS, Singla A. Drug resistance-associated genotypic alterations in the pol gene of HIV type 1 isolates in ART-naive individuals in North India. AIDS Res Hum Retroviruses. 2008;24:125–130. doi: 10.1089/aid.2007.0156. [DOI] [PubMed] [Google Scholar]
- 88.Deshpande A, Recordon-Pinson P, Deshmukh R, et al. Molecular characterization of HIV type 1 isolates from untreated patients of Mumbai (Bombay), India, and detection of rare resistance mutations. AIDS Res Hum Retroviruses. 2004;20:1032–1035. doi: 10.1089/aid.2004.20.1032. [DOI] [PubMed] [Google Scholar]
- 89.Eshleman SH, Hudelson SE, Gupta A, et al. Limited evolution in the HIV type 1 pol region among acute seroconverters in Pune, India. AIDS Res Hum Retroviruses. 2005;21:93–97. doi: 10.1089/aid.2005.21.93. [DOI] [PubMed] [Google Scholar]
- 90.Kandathil AJ, Kannangai R, Abraham OC, Sudarsanam TD, Pulimood SA, Sridharan G. Genotypic resistance profile of HIV-1 protease gene: a preliminary report from Vellore, south India. Indian J Med Microbiol. 2008;26:151–154. doi: 10.4103/0255-0857.40530. [DOI] [PubMed] [Google Scholar]
- 91.Lall M, Gupta RM, Sen S, Kapila K, Tripathy SP, Paranjape RS. Profile of primary resistance in HIV-1-infected treatment-naive individuals from Western India. AIDS Res Hum Retroviruses. 2008;24:987–990. doi: 10.1089/aid.2008.0079. [DOI] [PubMed] [Google Scholar]
- 92.Sen S, Tripathy SP, Chimanpure VM, Patil AA, Bagul RD, Paranjape RS. Human immunodeficiency virus type 1 drug resistance mutations in peripheral blood mononuclear cell proviral DNA among antiretroviral treatment-naive and treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007;23:489–497. doi: 10.1089/aid.2006.0221. [DOI] [PubMed] [Google Scholar]
- 93.Soheilli ZS, Ataiee Z, Tootian S, et al. Presence of HIV-1 CRF35_AD in Iran. AIDS Res Hum Retroviruses. 2009;25:123–124. doi: 10.1089/aid.2008.0199. [DOI] [PubMed] [Google Scholar]
- 94.Soundararajan L, Karunaianandham R, Jauvin V, et al. Characterization of HIV-1 isolates from antiretroviral drug-naive children in southern India. AIDS Res Hum Retroviruses. 2007;23:1119–1126. doi: 10.1089/aid.2007.0012. [DOI] [PubMed] [Google Scholar]
- 95.Balakrishnan P, Kumarasamy N, Kantor R, et al. HIV type 1 genotypic variation in an antiretroviral treatment-naive population in southern India. AIDS Res Hum Retroviruses. 2005;21:301–305. doi: 10.1089/aid.2005.21.301. [DOI] [PubMed] [Google Scholar]
- 96.Pandrea I, Descamps D, Collin G, et al. HIV type 1 genetic diversity and genotypic drug susceptibility in the Republic of Moldova. AIDS Res Hum Retroviruses. 2001;17:1297–1304. doi: 10.1089/088922201750461375. [DOI] [PubMed] [Google Scholar]
- 97.Zarandia M, Tsertsvadze T, Carr JK, Nadai Y, Sanchez JL, Nelson AK. HIV-1 genetic diversity and genotypic drug susceptibility in the Republic of Georgia. AIDS Res Hum Retroviruses. 2006;22:470–476. doi: 10.1089/aid.2006.22.470. [DOI] [PubMed] [Google Scholar]
- 98.Kolupajeva T, Aldins P, Guseva L, et al. HIV drug resistance tendencies in Latvia. Cent Eur J Public Health. 2008;16:138–140. doi: 10.21101/cejph.a3473. [DOI] [PubMed] [Google Scholar]
- 99.Vazquez de Parga E, Rakhmanova A, Perez-Alvarez L, et al. Analysis of drug resistance-associated mutations in treatment-naive individuals infected with different genetic forms of HIV-1 circulating in countries of the former Soviet Union. J Med Virol. 2005;77:337–344. doi: 10.1002/jmv.20461. [DOI] [PubMed] [Google Scholar]
- 100.Rodrigues R, Scherer LC, Oliveira CM, et al. Low prevalence of primary antiretroviral resistance mutations and predominance of HIV-1 clade C at polymerase gene in newly diagnosed individuals from south Brazil. Virus Res. 2006;116:201–207. doi: 10.1016/j.virusres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 101.Sa-Ferreira JA, Brindeiro PA, Chequer-Fernandez S, Tanuri A, Morgado MG. Human immunodeficiency virus-1 subtypes and antiretroviral drug resistance profiles among drug-naive Brazilian blood donors. Transfusion. 2007;47:97–102. doi: 10.1111/j.1537-2995.2007.01069.x. [DOI] [PubMed] [Google Scholar]
- 102.Soares EA, Santos RP, Pellegrini JA, Sprinz E, Tanuri A, Soares MA. Epidemiologic and molecular characterization of human immunodeficiency virus type 1 in southern Brazil. J Acquir Immune Defic Syndr. 2003;34:520–526. doi: 10.1097/00126334-200312150-00012. [DOI] [PubMed] [Google Scholar]
- 103.Perez L, Alvarez LP, Carmona R, et al. Genotypic resistance to antiretroviral drugs in patients infected with several HIV type 1 genetic forms in Cuba. AIDS Res Hum Retroviruses. 2007;23:407–414. doi: 10.1089/aid.2006.0155. [DOI] [PubMed] [Google Scholar]
-
104.Dilernia DA, Lourtau L, Gomez AM, et al. Drug-resistance surveillance among newly HIV-1 diagnosed individuals in Buenos Aires, Argentina. AIDS. 2007;21:1355–1360. doi: 10.1097/QAD.0b013e3280b07db1. [DOI] [PubMed] [Google Scholar];
 Well-designed study based on the WHO criteria for determining TDR in a selected population.
Well-designed study based on the WHO criteria for determining TDR in a selected population.
- 105.Petroni A, Deluchi G, Pryluka D, et al. Update on primary HIV-1 resistance in Argentina: emergence of mutations conferring high-level resistance to nonnucleoside reverse transcriptase inhibitors in drug-naive patients. J Acquir Immune Defic Syndr. 2006;42:506–510. doi: 10.1097/01.qai.0000222285.44460.e2. [DOI] [PubMed] [Google Scholar]
- 106.Barreto CC, Nishyia A, Araujo LV, Ferreira JE, Busch MP, Sabino EC. Trends in antiretroviral drug resistance and clade distributions among HIV-1-infected blood donors in Sao Paulo, Brazil. J Acquir Immune Defic Syndr. 2006;41:338–341. doi: 10.1097/01.qai.0000199097.88344.50. [DOI] [PubMed] [Google Scholar]
- 107.Paraschiv S, Otelea D, Dinu M, Maxim D, Tinischi M. Polymorphisms and resistance mutations in the protease and reverse transcriptase genes of HIV-1 F subtype Romanian strains. Int J Infect Dis. 2007;11:123–128. doi: 10.1016/j.ijid.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 108.Booth CL, Garcia-Diaz AM, Youle MS, Johnson MA, Phillips A, Geretti AM. Prevalence and predictors of antiretroviral drug resistance in newly diagnosed HIV-1 infection. J Antimicrob Chemother. 2007;59:517–524. doi: 10.1093/jac/dkl501. [DOI] [PubMed] [Google Scholar]
- 109.Jorgensen LB, Christensen MB, Gerstoft J, et al. Prevalence of drug resistance mutations and non-B subtypes in newly diagnosed HIV-1 patients in Denmark. Scand J Infect Dis. 2003;35:800–807. doi: 10.1080/00365540310016916. [DOI] [PubMed] [Google Scholar]
- 110.Kousiappa I, van de Vijver DA, Demetriades I, Kostrikis LG. Genetic analysis of HIV type 1 strains from newly infected untreated patients in cyprus: high genetic diversity and low prevalence of drug resistance. AIDS Res Hum Retroviruses. 2009;25:23–35. doi: 10.1089/aid.2008.0168. [DOI] [PubMed] [Google Scholar]
- 111.Vercauteren J, Derdelinckx I, Sasse A, et al. Prevalence and epidemiology of HIV type 1 drug resistance among newly diagnosed therapy-naive patients in Belgium from 2003 to 2006. AIDS Res Hum Retroviruses. 2008;24:355–362. doi: 10.1089/aid.2007.0212. [DOI] [PubMed] [Google Scholar]
- 112.Abreu CM, Brindeiro PA, Martins AN, et al. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 isolates circulating in pregnant women from Mozambique. Arch Virol. 2008;153:2013–2017. doi: 10.1007/s00705-008-0215-6. [DOI] [PubMed] [Google Scholar]
- 113.Adje-Toure C, Bile CE, Borget MY, et al. Polymorphism in protease and reverse transcriptase and phenotypic drug resistance of HIV-1 recombinant CRF02_AG isolates from patients with no prior use of antiretroviral drugs in Abidjan, Cote d'Ivoire. J Acquir Immune Defic Syndr. 2003;34:111–113. doi: 10.1097/00126334-200309010-00016. [DOI] [PubMed] [Google Scholar]
- 114.Agwale SM, Zeh C, Paxinos E, et al. Genotypic and phenotypic analyses of human immunodeficiency virus type 1 in antiretroviral drug-naive Nigerian patients. AIDS Res Hum Retroviruses. 2006;22:22–26. doi: 10.1089/aid.2006.22.22. [DOI] [PubMed] [Google Scholar]
- 115.Bellocchi MC, Forbici F, Palombi L, et al. Subtype analysis and mutations to antiviral drugs in HIV-1-infected patients from Mozambique before initiation of antiretroviral therapy: results from the DREAM programme. J Med Virol. 2005;76:452–458. doi: 10.1002/jmv.20382. [DOI] [PubMed] [Google Scholar]
- 116.Bessong PO, Mphahlele J, Choge IA, et al. Resistance mutational analysis of HIV type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses. 2006;22:1306–1312. doi: 10.1089/aid.2006.22.1306. [DOI] [PubMed] [Google Scholar]
- 117.Deho L, Walwema R, Cappelletti A, et al. Subtype assignment and phylogenetic analysis of HIV type 1 strains in patients from Swaziland. AIDS Res Hum Retroviruses. 2008;24:323–325. doi: 10.1089/aid.2007.0233. [DOI] [PubMed] [Google Scholar]
- 118.Fleury H, Recordon-Pinson P, Caumont A, et al. HIV type 1 diversity in France, 1999–2001: molecular characterization of non-B HIV type 1 subtypes and potential impact on susceptibility to antiretroviral drugs. AIDS Res Hum Retroviruses. 2003;19:41–47. doi: 10.1089/08892220360473952. [DOI] [PubMed] [Google Scholar]
- 119.Fonjungo PN, Mpoudi EN, Torimiro JN, et al. Human immunodeficiency virus type 1 group m protease in cameroon: genetic diversity and protease inhibitor mutational features. J Clin Microbiol. 2002;40:837–845. doi: 10.1128/JCM.40.3.837-845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gatanaga H, Ibe S, Matsuda M, et al. Drug-resistant HIV-1 prevalence in patients newly diagnosed with HIV/AIDS in Japan. Antiviral Res. 2007;75:75–82. doi: 10.1016/j.antiviral.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 121.Giuliani M, Montieri S, Palamara G, et al. Non-B HIV type 1 subtypes among men who have sex with men in Rome, Italy. AIDS Res Hum Retroviruses. 2009;25:157–164. doi: 10.1089/aid.2008.0175. [DOI] [PubMed] [Google Scholar]
- 122.Handema R, Terunuma H, Kasolo F, et al. Prevalence of drug-resistance-associated mutations in antiretroviral drug-naive Zambians infected with subtype C HIV-1. AIDS Res Hum Retroviruses. 2003;19:151–160. doi: 10.1089/088922203762688667. [DOI] [PubMed] [Google Scholar]
- 123.Jacobs GB, Laten A, van Rensburg EJ, et al. Phylogenetic diversity and low level antiretroviral resistance mutations in HIV type 1 treatment-naive patients from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24:1009–1012. doi: 10.1089/aid.2008.0028. [DOI] [PubMed] [Google Scholar]
- 124.Kassu A, Fujino M, Matsuda M, Nishizawa M, Ota F, Sugiura W. Molecular epidemiology of HIV type 1 in treatment-naive patients in north Ethiopia. AIDS Res Hum Retroviruses. 2007;23:564–568. doi: 10.1089/aid.2006.0270. [DOI] [PubMed] [Google Scholar]
- 125.Kinomoto M, Appiah-Opong R, Brandful JA, et al. HIV-1 proteases from drug-naive West African patients are differentially less susceptible to protease inhibitors. Clin Infect Dis. 2005;41:243–251. doi: 10.1086/431197. [DOI] [PubMed] [Google Scholar]
- 126.Konings FA, Zhong P, Agwara M, et al. Protease mutations in HIV-1 non-B strains infecting drug-naive villagers in Cameroon. AIDS Res Hum Retroviruses. 2004;20:105–109. doi: 10.1089/088922204322749558. [DOI] [PubMed] [Google Scholar]
- 127.Pandrea I, Robertson DL, Onanga R, et al. Analysis of partial pol and env sequences indicates a high prevalence of HIV type 1 recombinant strains circulating in Gabon. AIDS Res Hum Retroviruses. 2002;18:1103–1116. doi: 10.1089/088922202320567842. [DOI] [PubMed] [Google Scholar]
- 128.Sukasem C, Churdboonchart V, Sukeepaisarncharoen W, et al. Genotypic resistance profiles in antiretroviral-naive HIV-1 infections before and after initiation of first-line HAART: impact of polymorphism on resistance to therapy. Int J Antimicrob Agents. 2008;31:277–281. doi: 10.1016/j.ijantimicag.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 129.Toni T, Masquelier B, Bonard D, et al. Primary HIV-1 drug resistance in Abidjan (Cote d'Ivoire): a genotypic and phenotypic study. AIDS. 2002;16:488–491. doi: 10.1097/00002030-200202150-00024. [DOI] [PubMed] [Google Scholar]
- 130.Bocket L, Yazdanpanah Y, Ajana F, et al. Thymidine analogue mutations in antiretroviral-naive HIV-1 patients on triple therapy including either zidovudine or stavudine. J Antimicrob Chemother. 2004;53:89–94. doi: 10.1093/jac/dkh006. [DOI] [PubMed] [Google Scholar]
- 131.Frater AJ, Beardall A, Ariyoshi K, et al. Impact of baseline polymorphisms in RT and protease on outcome of highly active antiretroviral therapy in HIV-1-infected African patients. AIDS. 2001;15:1493–1502. doi: 10.1097/00002030-200108170-00006. [DOI] [PubMed] [Google Scholar]
- 132.Laurent C, Kouanfack C, Koulla-Shiro S, et al. Effectiveness and safety of a generic fixed-dose combination of nevirapine, stavudine, and lamivudine in HIV-1-infected adults in Cameroon: open-label multicentre trial. Lancet. 2004;364:29–34. doi: 10.1016/S0140-6736(04)16586-0. [DOI] [PubMed] [Google Scholar]
- 133.Tanuri A, Vicente AC, Otsuki K, et al. Genetic variation and susceptibilities to protease inhibitors among subtype B and F isolates in Brazil. Antimicrob Agents Chemother. 1999;43:253–258. doi: 10.1128/aac.43.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weidle PJ, Kityo CM, Mugyenyi P, et al. Resistance to antiretroviral therapy among patients in Uganda. J Acquir Immune Defic Syndr. 2001;26:495–500. doi: 10.1097/00126334-200104150-00017. [DOI] [PubMed] [Google Scholar]
- 135.Apetrei C, Descamps D, Collin G, et al. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J Virol. 1998;72:3534–3538. doi: 10.1128/jvi.72.5.3534-3538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van de Vijver DA, Wensing AM, Angarano G, et al. The calculated genetic barrier for antiretroviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J Acquir Immune Defic Syndr. 2006;41:352–360. doi: 10.1097/01.qai.0000209899.05126.e4. [DOI] [PubMed] [Google Scholar]
- 137.Blower SM, Aschenbach AN, Gershengorn HB, Kahn JO. Predicting the unpredictable: transmission of drug-resistant HIV. Nat Med. 2001;7:1016–1020. doi: 10.1038/nm0901-1016. [DOI] [PubMed] [Google Scholar]
- 138.Blower SM, Aschenbach AN, Kahn JO. Predicting the transmission of drug-resistant HIV: comparing theory with data. Lancet Infect Dis. 2003;3:10–11. doi: 10.1016/s1473-3099(03)00479-1. [DOI] [PubMed] [Google Scholar]
- 139.Vardavas R, Blower S. The emergence of HIV transmitted resistance in Botswana: “when will the WHO detection threshold be exceeded?”. PLoS ONE. 2007;2:e152. doi: 10.1371/journal.pone.0000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Blower S, Bodine E, Kahn J, McFarland W. The antiretroviral rollout and drug-resistant HIV in Africa: insights from empirical data and theoretical models. AIDS. 2005;19:1–14. doi: 10.1097/00002030-200501030-00001. [DOI] [PubMed] [Google Scholar]
- 141.Brenner BG, Roger M, Moisi DD, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22:2509–2515. doi: 10.1097/QAD.0b013e3283121c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Carrion G, Eyzaguirre L, Montano SM, et al. Documentation of subtype C HIV Type 1 strains in Argentina, Paraguay, and Uruguay. AIDS Res Hum Retroviruses. 2004;20:1022–1025. doi: 10.1089/aid.2004.20.1022. [DOI] [PubMed] [Google Scholar]
- 143.Delgado E, Leon-Ponte M, Villahermosa ML, et al. Analysis of HIV type 1 protease and reverse transcriptase sequences from Venezuela for drug resistance-associated mutations and subtype classification: a UNAIDS study. AIDS Res Hum Retroviruses. 2001;17:753–758. doi: 10.1089/088922201750237022. [DOI] [PubMed] [Google Scholar]
- 144.Ibe S, Hattori J, Fujisaki S, et al. Trend of drug-resistant HIV type 1 emergence among therapy-naive patients in Nagoya, Japan: an 8-year surveillance from 1999 to 2006. AIDS Res Hum Retroviruses. 2008;24:7–14. doi: 10.1089/aid.2007.0129. [DOI] [PubMed] [Google Scholar]
- 145.Kim CO, Chin BS, Han SH, et al. Low prevalence of drug-resistant HIV-1 in patients newly diagnosed with early stage of HIV infection in Korea. Tohoku J Exp Med. 2008;216:259–265. doi: 10.1620/tjem.216.259. [DOI] [PubMed] [Google Scholar]
- 146.Kojima Y, Kawahata T, Mori H, Oishi I, Otake T. Recent diversity of human immunodeficiency virus type 1 in individuals who visited sexually transmitted infection-related clinics in Osaka, Japan. J Infect Chemother. 2008;14:51–55. doi: 10.1007/s10156-007-0579-7. [DOI] [PubMed] [Google Scholar]
- 147.Lloyd B, O'Connell RJ, Michael NL, et al. Prevalence of resistance mutations in HIV-1-Infected Hondurans at the beginning of the National Antiretroviral Therapy Program. AIDS Res Hum Retroviruses. 2008;24:529–535. doi: 10.1089/aid.2007.0172. [DOI] [PubMed] [Google Scholar]
- 148.Tossonian HK, Raffa JD, Grebely J, et al. Primary drug resistance in antiretroviral-naive injection drug users. Int J Infect Dis. 2008 doi: 10.1016/j.ijid.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 149.Valle-Bahena OM, Ramos-Jimenez J, Ortiz-Lopez R, et al. Frequency of protease and reverse transcriptase drug resistance mutations in naive HIV-infected patients. Arch Med Res. 2006;37:1022–1027. doi: 10.1016/j.arcmed.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 150.Yilmaz G, Midilli K, Turkoglu S, et al. Genetic subtypes of human immunodeficiency virus type 1 (HIV-1) in Istanbul, Turkey. Int J Infect Dis. 2006;10:286–290. doi: 10.1016/j.ijid.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 151.Vergne L, Peeters M, Mpoudi-Ngole E, et al. Genetic diversity of protease and reverse transcriptase sequences in nonsubtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol. 2000;38:3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vergne L, Stuyver L, van Houtte M, Butel C, Delaporte E, Peeters M. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J Clin Virol. 2006;36:43–49. doi: 10.1016/j.jcv.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 153.Bussmann H, Novitsky V, Wester W, et al. HIV-1 subtype C drug-resistance background among ARV-naive adults in Botswana. Antivir Chem Chemother. 2005;16:103–115. doi: 10.1177/095632020501600203. [DOI] [PubMed] [Google Scholar]
- 154.Nadembega WM, Giannella S, Simpore J, et al. Characterization of drug-resistance mutations in HIV-1 isolates from non-HAART and HAART treated patients in Burkina Faso. J Med Virol. 2006;78:1385–1391. doi: 10.1002/jmv.20709. [DOI] [PubMed] [Google Scholar]
- 155.Tebit DM, Ganame J, Sathiandee K, Nagabila Y, Coulibaly B, Krausslich HG. Diversity of HIV in rural Burkina Faso. J Acquir Immune Defic Syndr. 2006;43:144–152. doi: 10.1097/01.qai.0000228148.40539.d3. [DOI] [PubMed] [Google Scholar]
- 156.Vergne L, Diagbouga S, Kouanfack C, et al. HIV-1 drug-resistance mutations among newly diagnosed patients before scaling-up programmes in Burkina Faso and Cameroon. Antivir Ther. 2006;11:575–579. [PubMed] [Google Scholar]
- 157.Vidal N, Niyongabo T, Nduwimana J, et al. HIV type 1 diversity and antiretroviral drug resistance mutations in Burundi. AIDS Res Hum Retroviruses. 2007;23:175–180. doi: 10.1089/aid.2006.0126. [DOI] [PubMed] [Google Scholar]
- 158.Koizumi Y, Ndembi N, Miyashita M, et al. Emergence of antiretroviral therapy resistance-associated primary mutations among drug-naive HIV-1-infected individuals in rural western Cameroon. J Acquir Immune Defic Syndr. 2006;43:15–22. doi: 10.1097/01.qai.0000226793.16216.55. [DOI] [PubMed] [Google Scholar]
- 159.McNulty A, Jennings C, Bennett D, et al. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J Clin Microbiol. 2007;45:517–521. doi: 10.1128/JCM.02016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ndembi N, Abraha A, Pilch H, et al. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol. 2008;46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ndongmo CB, Pieniazek D, Holberg-Petersen M, et al. HIV genetic diversity in Cameroon: possible public health importance. AIDS Res Hum Retroviruses. 2006;22:812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- 162.Tebit DM, Zekeng L, Kaptue L, Salminen M, Krausslich HG, Herchenroder O. Genotypic and phenotypic analysis of HIV type 1 primary isolates from western Cameroon. AIDS Res Hum Retroviruses. 2002;18:39–48. doi: 10.1089/088922202753394709. [DOI] [PubMed] [Google Scholar]
- 163.Vessiere A, Nerrienet E, Kfutwah A, et al. HIV-1 pol gene polymorphism and antiretroviral resistance mutations in drug-naive pregnant women in Yaounde, Cameroon. J Acquir Immune Defic Syndr. 2006;42:256–258. doi: 10.1097/01.qai.0000209909.20373.3b. [DOI] [PubMed] [Google Scholar]
- 164.Marechal V, Jauvin V, Selekon B, et al. Increasing HIV type 1 polymorphic diversity but no resistance to antiretroviral drugs in untreated patients from Central African Republic: a 2005 study. AIDS Res Hum Retroviruses. 2006;22:1036–1044. doi: 10.1089/aid.2006.22.1036. [DOI] [PubMed] [Google Scholar]
- 165.Punzi G, Saracino A, Brindicci G, et al. HIV infection and protease genetic diversity in a rural area of the Southern Central African Republic. J Med Virol. 2005;77:457–459. doi: 10.1002/jmv.20476. [DOI] [PubMed] [Google Scholar]
- 166.Toni TD, Recordon-Pinson P, Minga A, et al. Presence of key drug resistance mutations in isolates from untreated patients of Abidjan, Cote d'Ivoire: ANRS 1257 study. AIDS Res Hum Retroviruses. 2003;19:713–717. doi: 10.1089/088922203322280946. [DOI] [PubMed] [Google Scholar]
- 167.Vidal N, Mulanga C, Bazepeo SE, et al. HIV type 1 pol gene diversity and antiretroviral drug resistance mutations in the Democratic Republic of Congo (DRC) AIDS Res Hum Retroviruses. 2006;22:202–206. doi: 10.1089/aid.2006.22.202. [DOI] [PubMed] [Google Scholar]
- 168.Loemba H, Brenner B, Parniak MA, et al. Polymorphisms of cytotoxic T-lymphocyte (CTL) and T-helper epitopes within reverse transcriptase (RT) of HIV-1 subtype C from Ethiopia and Botswana following selection of antiretroviral drug resistance. Antiviral Res. 2002;56:129–142. doi: 10.1016/s0166-3542(02)00100-6. [DOI] [PubMed] [Google Scholar]
- 169.Caron M, Makuwa M, Souquiere S, Descamps D, Brun-Vezinet F, Kazanji M. Human immunodeficiency virus type 1 seroprevalence and antiretroviral drug resistance-associated mutations in miners in Gabon, central Africa. AIDS Res Hum Retroviruses. 2008;24:1225–1228. doi: 10.1089/aid.2008.0097. [DOI] [PubMed] [Google Scholar]
- 170.Vergne L, Malonga-Mouellet G, Mistoul I, et al. Resistance to antiretroviral treatment in Gabon: need for implementation of guidelines on antiretroviral therapy use and HIV-1 drug resistance monitoring in developing countries. J Acquir Immune Defic Syndr. 2002;29:165–168. doi: 10.1097/00042560-200202010-00009. [DOI] [PubMed] [Google Scholar]
- 171.Sagoe KW, Dwidar M, Lartey M, et al. Variability of the human immunodeficiency virus type 1 polymerase gene from treatment naive patients in Accra, Ghana. J Clin Virol. 2007;40:163–167. doi: 10.1016/j.jcv.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 172.Dowling WE, Kim B, Mason CJ, et al. Forty-one near full-length HIV-1 sequences from Kenya reveal an epidemic of subtype A and A-containing recombinants. AIDS. 2002;16:1809–1820. doi: 10.1097/00002030-200209060-00015. [DOI] [PubMed] [Google Scholar]
- 173.Yang C, Li M, Shi YP, et al. Genetic diversity and high proportion of intersubtype recombinants among HIV type 1-infected pregnant women in Kisumu, western Kenya. AIDS Res Hum Retroviruses. 2004;20:565–574. doi: 10.1089/088922204323087822. [DOI] [PubMed] [Google Scholar]
- 174.Razafindratsimandresy R, Rajaonatahina D, Soares JL, Rousset D, Reynes JM. High HIV type 1 subtype diversity and few drug resistance mutations among seropositive people detected during the 2005 second generation HIV surveillance in Madagascar. AIDS Res Hum Retroviruses. 2006;22:595–597. doi: 10.1089/aid.2006.22.595. [DOI] [PubMed] [Google Scholar]
- 175.Petch LA, Hoffman IF, Jere CS, et al. Genotypic analysis of the protease and reverse transcriptase of HIV type 1 subtype C isolates from antiretroviral drug-naive adults in Malawi. AIDS Res Hum Retroviruses. 2005;21:799–805. doi: 10.1089/aid.2005.21.799. [DOI] [PubMed] [Google Scholar]
- 176.Derache A, Maiga AI, Traore O, et al. Evolution of genetic diversity and drug resistance mutations in HIV-1 among untreated patients from Mali between 2005 and 2006. J Antimicrob Chemother. 2008;62:456–463. doi: 10.1093/jac/dkn234. [DOI] [PubMed] [Google Scholar]
- 177.Derache A, Traore O, Koita V, et al. Genetic diversity and drug resistance mutations in HIV type 1 from untreated patients in Bamako, Mali. Antivir Ther. 2007;12:123–129. [PubMed] [Google Scholar]
- 178.Lahuerta M, Aparicio E, Bardaji A, et al. Rapid spread and genetic diversification of HIV type 1 subtype C in a rural area of southern Mozambique. AIDS Res Hum Retroviruses. 2008;24:327–335. doi: 10.1089/aid.2007.0134. [DOI] [PubMed] [Google Scholar]
- 179.Ojesina AI, Sankale JL, Odaibo G, et al. Subtype-specific patterns in HIV Type 1 reverse transcriptase and protease in Oyo State, Nigeria: implications for drug resistance and host response. AIDS Res Hum Retroviruses. 2006;22:770–779. doi: 10.1089/aid.2006.22.770. [DOI] [PubMed] [Google Scholar]
- 180.Servais J, Lambert C, Karita E, et al. HIV type 1 pol gene diversity and archived nevirapine resistance mutation in pregnant women in Rwanda. AIDS Res Hum Retroviruses. 2004;20:279–283. doi: 10.1089/088922204322996518. [DOI] [PubMed] [Google Scholar]
- 181.Vergne L, Kane CT, Laurent C, et al. Low rate of genotypic HIV-1 drug-resistant strains in the Senegalese government initiative of access to antiretroviral therapy. AIDS. 2003;17(Suppl 3):S31–SS8. doi: 10.1097/00002030-200317003-00005. [DOI] [PubMed] [Google Scholar]
- 182.Razafindratsimandresy R, Hollanda J, Soares JL, Rousset D, Chetty AP, Reynes JM. HIV type 1 diversity in the Seychelles. AIDS Res Hum Retroviruses. 2007;23:761–763. doi: 10.1089/aid.2007.0002. [DOI] [PubMed] [Google Scholar]
- 183.Maphalala G, Okello V, Mndzebele S, et al. Surveillance of transmitted HIV drug resistance in the Manzini-Mbabane corridor, Swaziland, in 2006. Antivir Ther. 2008;13(Suppl 2):95–100. [PubMed] [Google Scholar]
- 184.Arroyo MA, Hoelscher M, Sateren W, et al. HIV-1 diversity and prevalence differ between urban and rural areas in the Mbeya region of Tanzania. AIDS. 2005;19:1517–1524. doi: 10.1097/01.aids.0000183515.14642.76. [DOI] [PubMed] [Google Scholar]
- 185.Arroyo MA, Hoelscher M, Sanders-Buell E, et al. HIV type 1 subtypes among blood donors in the Mbeya region of southwest Tanzania. AIDS Res Hum Retroviruses. 2004;20:895–901. doi: 10.1089/0889222041725235. [DOI] [PubMed] [Google Scholar]
- 186.Nyombi BM, Kristiansen KI, Bjune G, Muller F, Holm-Hansen C. Diversity of human immunodeficiency virus type 1 subtypes in Kagera and Kilimanjaro regions, Tanzania. AIDS Res Hum Retroviruses. 2008;24:761–769. doi: 10.1089/aid.2007.0311. [DOI] [PubMed] [Google Scholar]
- 187.Somi GR, Kibuka T, Diallo K, et al. Surveillance of transmitted HIV drug resistance among women attending antenatal clinics in Dar es Salaam, Tanzania. Antivir Ther. 2008;13(Suppl 2):77–82. [PubMed] [Google Scholar]
- 188.Becker-Pergola G, Kataaha P, Johnston-Dow L, Fung S, Jackson JB, Eshleman SH. Analysis of HIV type 1 protease and reverse transcriptase in antiretroviral drug-naive Ugandan adults. AIDS Res Hum Retroviruses. 2000;16:807–813. doi: 10.1089/088922200308800. [DOI] [PubMed] [Google Scholar]
- 189.Church JD, Hudelson SE, Guay LA, et al. HIV type 1 variants with nevirapine resistance mutations are rarely detected in antiretroviral drug-naive African women with subtypes A, C, and D. AIDS Res Hum Retroviruses. 2007;23:764–768. doi: 10.1089/aid.2006.0272. [DOI] [PubMed] [Google Scholar]
- 190.Gale CV, Yirrell DL, Campbell E, van der Paal L, Grosskurth H, Kaleebu P. Genotypic variation in the pol gene of HIV type 1 in an antiretroviral treatment-naive population in rural southwestern Uganda. AIDS Res Hum Retroviruses. 2006;22:985–992. doi: 10.1089/aid.2006.22.985. [DOI] [PubMed] [Google Scholar]
- 191.Ndembi N, Lyagoba F, Nanteza B, et al. Transmitted antiretroviral drug resistance surveillance among newly HIV type 1-diagnosed women attending an antenatal clinic in Entebbe, Uganda. AIDS Res Hum Retroviruses. 2008;24:889–895. doi: 10.1089/aid.2007.0317. [DOI] [PubMed] [Google Scholar]
- 192.Shafer RW, Chuang TK, Hsu P, White CB, Katzenstein DA. Sequence and drug susceptibility of subtype C protease from human immunodeficiency virus type 1 seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 1999;15:65–69. doi: 10.1089/088922299311727. [DOI] [PubMed] [Google Scholar]
- 193.Saad MD, Aliev Q, Botros BA, et al. Genetic forms of HIV Type 1 in the former Soviet Union dominate the epidemic in Azerbaijan. AIDS Res Hum Retroviruses. 2006;22:796–800. doi: 10.1089/aid.2006.22.796. [DOI] [PubMed] [Google Scholar]
- 194.Hai-Long H, Jian Z, Ping-Ping Y, et al. Genetic characterization of CRF01_AE full-length human immunodeficiency virus type 1 sequences from Fujian, China. AIDS Res Hum Retroviruses. 2007;23:569–574. doi: 10.1089/aid.2006.0257. [DOI] [PubMed] [Google Scholar]
- 195.Laeyendecker O, Zhang GW, Quinn TC, et al. Molecular epidemiology of HIV-1 subtypes in southern China. J Acquir Immune Defic Syndr. 2005;38:356–362. [PubMed] [Google Scholar]
- 196.Liu J, Yue J, Wu S, Yan Y. Polymorphisms and drug resistance analysis of HIV-1 CRF01_AE strains circulating in Fujian Province, China. Arch Virol. 2007;152:1799–1805. doi: 10.1007/s00705-007-1019-9. [DOI] [PubMed] [Google Scholar]
- 197.Grossman Z, Lorber M, Maayan S, et al. Drug-resistant HIV infection among drug-naive patients in Israel. Clin Infect Dis. 2005;40:294–302. doi: 10.1086/426592. [DOI] [PubMed] [Google Scholar]
- 198.Eyzaguirre LM, Erasilova IB, Nadai Y, et al. Genetic characterization of HIV-1 strains circulating in Kazakhstan. J Acquir Immune Defic Syndr. 2007;46:19–23. doi: 10.1097/QAI.0b013e318073c620. [DOI] [PubMed] [Google Scholar]
- 199.Tee KK, Pon CK, Kamarulzaman A, Ng KP. Emergence of HIV-1 CRF01_AE/B unique recombinant forms in Kuala Lumpur, Malaysia. AIDS. 2005;19:119–126. doi: 10.1097/00002030-200501280-00003. [DOI] [PubMed] [Google Scholar]
- 200.Roudinskii NI, Sukhanova AL, Kazennov EV, et al. Diversity of human immunodeficiency virus type 1 subtype A and CRF03_AB protease in eastern Europe: selection of the V77I variant and its rapid spread in injecting drug user populations. J Virol. 2004;78:11276–11287. doi: 10.1128/JVI.78.20.11276-11287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Saad MD, Shcherbinskaya AM, Nadai Y, et al. Molecular epidemiology of HIV type 1 in Ukraine: birthplace of an epidemic. AIDS Res Hum Retroviruses. 2006;22:709–714. doi: 10.1089/aid.2006.22.709. [DOI] [PubMed] [Google Scholar]
- 202.Carr JK, Nadai Y, Eyzaguirre L, et al. Outbreak of a West African recombinant of HIV-1 in Tashkent, Uzbekistan. J Acquir Immune Defic Syndr. 2005;39:570–575. [PubMed] [Google Scholar]
- 203.Caumont A, Lan NT, Uyen NT, et al. Sequence analysis of env C2/V3, gag p17/p24, and pol protease regions of 25 HIV type 1 isolates from Ho Chi Minh City, Vietnam. AIDS Res Hum Retroviruses. 2001;17:1285–1291. doi: 10.1089/088922201750461357. [DOI] [PubMed] [Google Scholar]
- 204.Lan NT, Recordon-Pinson P, Hung PV, et al. HIV type 1 isolates from 200 untreated individuals in Ho Chi Minh City (Vietnam): ANRS 1257 Study. Large predominance of CRF01_AE and presence of major resistance mutations to antiretroviral drugs. AIDS Res Hum Retroviruses. 2003;19:925–928. doi: 10.1089/088922203322493111. [DOI] [PubMed] [Google Scholar]
- 205.Saad MD, Al-Jaufy A, Grahan RR, et al. HIV type 1 strains common in Europe, Africa, and Asia cocirculate in Yemen. AIDS Res Hum Retroviruses. 2005;21:644–648. doi: 10.1089/aid.2005.21.644. [DOI] [PubMed] [Google Scholar]
- 206.Ciccozzi M, Gori C, Boros S, et al. Molecular diversity of HIV in Albania. J Infect Dis. 2005;192:475–479. doi: 10.1086/431599. [DOI] [PubMed] [Google Scholar]
- 207.Paraskevis D, Magiorkinis E, Katsoulidou A, et al. Prevalence of resistance-associated mutations in newly diagnosed HIV-1 patients in Greece. Virus Res. 2005;112:115–122. doi: 10.1016/j.virusres.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 208.Palma AC, Araujo F, Duque V, Borges F, Paixao MT, Camacho R. Molecular epidemiology and prevalence of drug resistance-associated mutations in newly diagnosed HIV-1 patients in Portugal. Infect Genet Evol. 2007;7:391–398. doi: 10.1016/j.meegid.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 209.Parreira R, Santos M, Piedade J, Esteves A. Natural polymorphism of HIV-1 subtype G protease and cleavage sites. AIDS. 2004;18:1345–1346. doi: 10.1097/00002030-200406180-00017. [DOI] [PubMed] [Google Scholar]
- 210.Babic DZ, Zelnikar M, Seme K, et al. Prevalence of antiretroviral drug resistance mutations and HIV-1 non-B subtypes in newly diagnosed drug-naive patients in Slovenia, 2000–2004. Virus Res. 2006;118:156–163. doi: 10.1016/j.virusres.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 211.Holguin A, Alvarez A, Soriano V. High prevalence of HIV-1 subtype G and natural polymorphisms at the protease gene among HIV-infected immigrants in Madrid. AIDS. 2002;16:1163–1170. doi: 10.1097/00002030-200205240-00010. [DOI] [PubMed] [Google Scholar]
- 212.Perez-Alvarez L, Carmona R, Munoz M, et al. High incidence of non-B and recombinant HIV-1 strains in newly diagnosed patients in Galicia, Spain: study of genotypic resistance. Antivir Ther. 2003;8:355–360. [PubMed] [Google Scholar]
- 213.Maljkovic I, Wilbe K, Solver E, Alaeus A, Leitner T. Limited transmission of drug-resistant HIV type 1 in 100 Swedish newly detected and drug-naive patients infected with subtypes A, B, C, D, G, U, and CRF01_AE. AIDS Res Hum Retroviruses. 2003;19:989–997. doi: 10.1089/088922203322588341. [DOI] [PubMed] [Google Scholar]
- 214.Holguin A, Paxinos E, Hertogs K, Womac C, Soriano V. Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitors in HIV-1 non-B subtypes. J Clin Virol. 2004;31:215–220. doi: 10.1016/j.jcv.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 215.Tovanabutra S, Brodine SK, Mascola JR, et al. Characterization of complete HIV type 1 genomes from non-B subtype infections in U.S. military personnel. AIDS Res Hum Retroviruses. 2005;21:424–429. doi: 10.1089/aid.2005.21.424. [DOI] [PubMed] [Google Scholar]
Websites
- 301.HIV Sequence Database. Breakpoints of circulating recombinant forms. [1 April 2009]; www.hiv.lanl.gov/content/sequence/HIV/CRFs/breakpoints.html.
- 302.French National Agency for AIDS Research, v17. [1 April 2009]; www.hivfrenchresistance.org.
- 303.Stanford Database. [1 April 2009]; http://hivdb.stanford.edu.
- 304.Rega Institute mutation list v8.0.1. [1 April 2009]; www.rega.kuleuven.be/cev/


