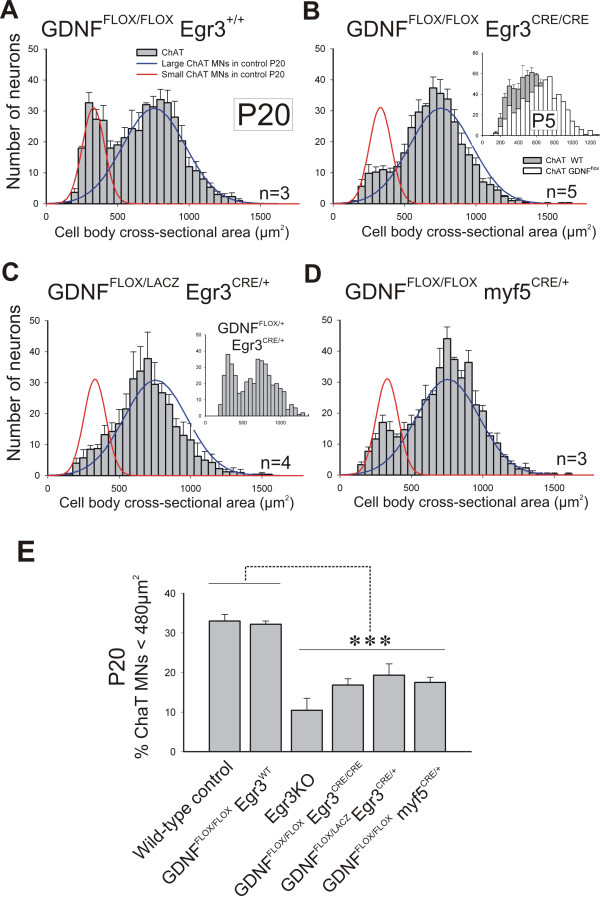
Figure 7.
Genetic elimination of muscle spindle-derived GDNF results in selective loss of gamma motor neurons. (A) Size distributions of ChAT+ MNs in GDNFFLOX/FLOX/Egr3WT (no Cre) controls at P20 are comparable to wild types (lines). (B) ChAT+ MNs losses in the absence of spindle-derived GDNF (GDNFFLOX/FLOX/Egr3CRE/CRE mutants). Small ChAT+ MNs represent 32 ± 1% ( ± SEM) of all MNs in GDNFFLOX/FLOX /Egr3+/+ and 16.8 ± 1.1% in GDNFFLOX/FLOX/Egr3CRE/CRE animals. Inset shows a depletion at P5 comparable to Egr3KO animals (see Figure 5F). (C) Similar loses in compound heterozygotes with one conditional and one null GDNF allele and a single copy of Egr3CRE (GDNFFLOX/LACZ/Egr3CRE/+). Inset shows a normal size distribution in one animal carrying one wild type and one floxed GDNF allele and a single Egr3CRE copy. (D) GDNF elimination from all muscle precursors using myf5CRE/+ results in similar losses of small ChAT+ MNs. Large MN numbers are unaffected in conditional mutants by targeted removal of GDNF from spindles. (E) Comparison of the percentage of small MNs (<480 μm2) in different genotypes. No differences were detected between wild-type and homozygous GDNFFLOX (no Cre) controls. Egr3KO mutants and several conditional/floxed GDNF mutants crossed to Egr3CRE or myf5CRE showed significant depletions compared to wild-types and GDNFFLOX (no Cre) controls (asterisks indicate P < 0.001 one-way ANOVA followed by P < 0.01 post-hoc Tukey comparisons). Depletion of small MNs in Egr3KO animals were more pronounced than in other genotypes, but differences were not statistically significant. N's, number of animals analyzed in each genotype.