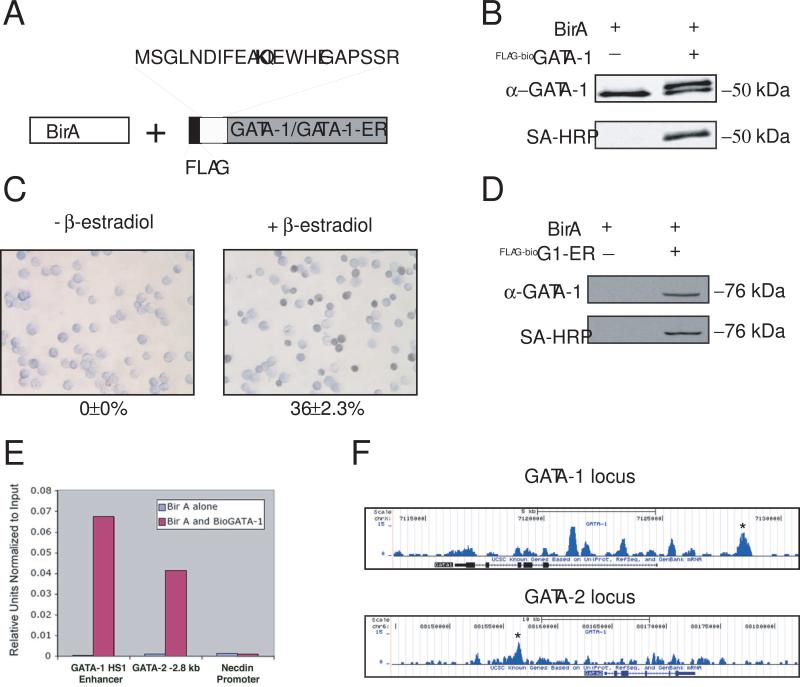
Figure 1.
Streptavidin ChIP-Seq of metabolically biotinylated GATA-1 in induced MEL cells. (A) Schematic diagram of metabolic biotin tagging system. birA, E.coli biotin ligase. The birA recognition motif and FLAG epitope tag is shown fused to the amino terminus of GATA-1 or GATA-1ER. The biotin acceptor lysine is indicated in bold. (B) Western blot of nuclear extracts from MEL cell clones expressing either birA alone, or birA and the recombinant GATA-1 (FLAG-BioGATA-1). Upper panel, probed with anti-GATA-1 antibody; Lower panel, same as upper panel after stripping and re-probing with streptavidin-horse radish peroxidase (SA-HRP). (C) O-dianosidine (benzidine) stain of G1E cells expressing FLAG-BioGATA-1-ER without (left) or with (right) treatment with β-estradiol for 48 hrs. Hemoglobinized cells stain dark brown/black. The percentage of positive cells is indicated below the panel (−/+ standard error of the mean (SEM); n=3). (D) Western blot of nuclear extracts from cell shown in “C” probed with anti-GATA-1 antibody (top) and SA-HRP (bottom). (E) Quantitative ChIP assay at two known GATA-1 occupancy sites (GATA-1 HS1 and GATA-2 “−2.8 kb” enhancer) and a negative control site (necdin promoter) using streptavidin-based ChIP from induced MEL cells (DMSO for 24 hrs) expressing birA alone or birA and FLAG-BioGATA-1. (F) Streptavidin ChIP-Seq enrichment profiles for FLAG-BioGATA-1 at the GATA-1 and GATA-2 loci. Enrichment peaks corresponding to GATA-1 HS1 and GATA-2 “−2.8 kb” are indicated with asterisks.