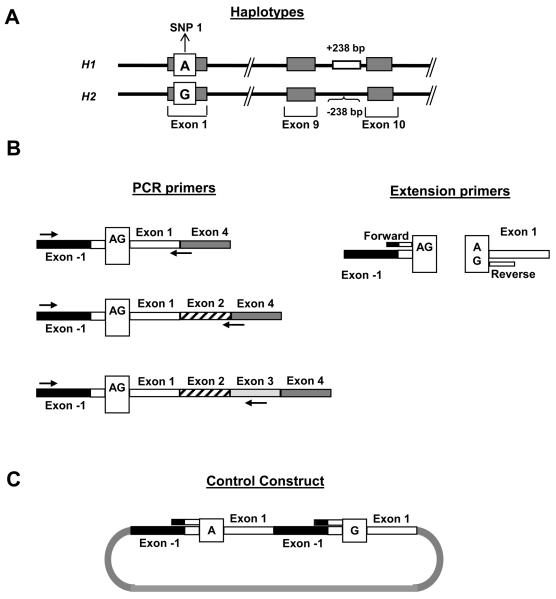
Figure 1.
Primer design for the MALDI-ToF MS assays for expression of MAPT N-terminal exons. The methodology requires a reverse transcription PCR (RT-PCR) reaction, followed by a primer extension reaction and analysis by MALDI-ToF mass spectrometry. (A) The haplotype tagging variant SNP 1, used to assay expression of the alternatively splice N-terminal exons, is in complete linkage disequilibrium with the haplotype defined 238 bp deletion located in intron 9. The SNP 1 alleles are indicated in the white box, with the H1 shown above the H2 allele. (B). The RT-PCR primers used to amplify fragments containing SNP 1 are shown on the left, and the locations of the extension primers used for the single base extension reaction extending to the SNP are shown on the right. Reverse RT-PCR primers were strategically placed to amplify either the 0N, 1N or 2N MAPT transcripts. On the right, the forward and reverse base-extension primers are indicated by short bars. (C) Structure of the artificial construct built for the forward base-extension assay at SNP 1 to normalize the MALDI-ToF data to the expected 1:1 ratio. The SNP 1 forward assay uses an extension primer that spans the exon −1/exon 1 boundary, a sequence that does not exist in genomic DNA.