Abstract
Strawberry anthracnose, caused by Colletotrichum spp., is a major disease of cultivated strawberry. This study identifies 31 isolates of Colletotrichum spp. which cause strawberry anthracnose in Zhejiang Province and Shanghai City, China. Eleven isolates were identified as C. acutatum, 10 as C. gloeosporioides and 10 as C. fragariae based on morphological characteristics, phylogenetic and sequence analyses. Species-specific polymerase chain reaction (PCR) and enzyme digestion further confirmed the identification of the Colletotrichum spp., demonstrating that these three species are currently the causal agents of strawberry anthracnose in the studied regions. Based on analysis of rDNA internal transcribed spacers (ITS) sequences, sequences of all C. acutatum were identical, and little genetic variability was observed between C. fragariae and C. gloeosporioides. However, the conservative nature of the MvnI specific site from isolates of C. gloeosporioides was confirmed, and this site could be used to differentiate C. gloeosporioides from C. fragariae.
Keywords: Fragaria×ananassa, Taxonomic identification, Colletotrichum acutatum, C. gloeosporioides, C. fragariae
1. Introduction
Colletotrichum species cause economically significant diseases including wilts, rots and anthracnose in production areas of strawberry (Fragaria×ananassa Duch.) worldwide (Buddie et al., 1999; Debode et al., 2009). Three species have been reported as causal agents of strawberry anthracnose: C. acutatum Simmonds (teleomorph Glomerella acutata J.C. Guerber & J.C. Correll), C. gloeosporioides (Penz.) Penz. & Sacc. in Penz. [teleomorph Glomerella cingulata (Stoneman) Spauld. & Schrenk], and C. fragariae Brooks (Gunnell and Gubler, 1992; Denoyes-Rothan et al., 2003). C. gloeosporioides and C. acutatum are distributed worldwide on a number of hosts, while C. fragariae has a very narrow host range (MacKenzie et al., 2008). The disease causes up to 80% of plant deaths in nurseries and over 50% of yield losses in strawberry production fields (Sreenivasaprasad and Talhinhas, 2005). Likewise, in the Zhejiang region, nearly 50% of seedling deaths in nurseries and over 40% of yield losses in strawberry production fields are caused by this disease according to our investigations.
Strawberry is cultivated extensively in some districts of China and severe levels of strawberry anthracnose have been prevalent for many years. Shao (1992) first observed strawberry anthracnose disease in Shanghai on cv. Lego introduced from Japan and the pathogen was identified as C. acutatum (Dai et al., 2006). Subsequently, Ye et al. (1997) reported that the pathogens responsible for strawberry anthracnose in Shanghai belonged to C. fragariae and C. gloeosporioides, based on pathogen morphological characteristics. However, based on a combination of morphology and rDNA internal transcribed spacers (ITS) sequence analysis, C. acutatum and C. gloeosporioides were identified as the causal agents (Ren et al., 2008). Zhejiang Province, adjacent to Shanghai, is an important strawberry-producing area with about 6 000 ha under cultivation. Although the disease occurs every year, isolation and molecular identification to clarify which Colletotrichum spp. are responsible for anthracnose disease in the area have not been performed. The main purpose of this study was to establish which species are currently causing anthracnose disease in strawberry in Zhejiang Province and to compare them with isolates from strawberry plants from Shanghai.
2. Materials and methods
2.1. Fungal isolates
Colletotrichum spp. were isolated from diseased strawberry plants (fruit, vegetative parts, and root crowns) from different cultivars and sites from the summer of 2006 to the spring of 2007. Infected plant organs were surface-sterilized [0.5% (w/v) sodium hypochlorite] for 2–5 min before they were placed onto potato dextrose agar (PDA) containing streptomycin. Isolates selected were transferred to PDA, and single conidial isolates were prepared from each isolate. Cultures were grown on PDA at 25 °C. Plugs (5 mm) were cut from the margin of fresh colonies and stored in 20% (w/v) glycerol in a freezer at −70 °C.
2.2. Morphological examination
Isolates representative of different sites were selected for morphological species identification. These isolates were cultured on PDA in darkness at 25 °C for 5 d. Mycelial discs (5 mm) were taken from the growing edge of colonies and transferred onto plates of strawberry leaf agar (SLA) (Gunnell and Gubler, 1992). The SLA plates were placed in an incubator under continuous fluorescent light at 25 °C. Conidia were harvested after 14 d, and setae after 22–30 d, from acervuli produced on the leaf pieces. Conidia were mounted in water, and examined for size and shape, using a Zeiss Axiophot 2 microscope with an Axiocam CCD camera and AxioVision digital imaging software (AxioVision Software Release 3.1, version 3-2002; Carl Zeiss Vision Imaging Systems). Colony color was determined after 7 d on PDA at 25 °C under conditions of alternating cycles of 12 h light and 12 h darkness.
2.3. DNA extraction
Fungal isolates were cultured in darkness at 25 °C for 5 d. Mycelial disks were excised from the margins of colonies and inoculated into 100 ml of liquid growth media potato dextrose broth (PDB) in 250 ml flasks, shaken at 200 r/min at 25 °C for 5 d. Mycelia were harvested by filtration, freeze-dried, ground to a fine powder in liquid nitrogen, and then stored at −70 °C. About 50 mg of mycelial powder was removed into a sterile 1.5-ml tube, rehydrated in 600 μl of 2× cetyltrimethylammonium bromide (CTAB) buffer [100 mmol/L Tris (pH 8.0), 1.4 mol/L NaCl, 30 mmol/L ethylenediaminetetraacetic acid (EDTA), 2% (w/v) hexadecyltrimethylammonium bromide], and incubated in a water bath at 65 °C for 30–60 min. Following a phenol/chloroform extraction, the genomic DNA was precipitated by isopropanol in the presence of sodium acetate. Genomic DNA was visualized in 1% (w/v) agarose gels after ethidium bromide staining. DNA concentration was determined by ultraviolet (UV) spectroscopy.
2.4. Amplification of rDNA-ITS regions
The ITS regions of rDNA were amplified using the primers ITS6 (5′-GAAGGTGAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) for direct sequencing (Cooke and Duncan, 1997). Polymerase chain reaction (PCR) amplifications were performed in a total volume of 50 μl containing 40 mmol/L of Tris-HCl (pH 8.4), 100 mmol/L of KCl, 3 mmol/L of MgCl2, 200 μmol/L of each dNTP, 1 μmol/L of each primer, and 0.5 U of Taq (Sangon Co., Shanghai, China). PCR amplifications were carried out using a PTC-150 MiniCycler (MJ Research Corp., America). Following an initial denaturation at 95 °C for 4 min, the DNA templates were amplified for 35 cycles. Each cycle consisted of a denaturation step at 95 °C for 1 min, an annealing step at 52 °C for 1 min, and an extension step at 72 °C for 1 min. A final extension step (72 °C for 10 min) was included at the end. Aliquots of 4 μl of the amplification products were electrophoresed on 1% (w/v) agarose gels, and the PCR products were subsequently stained with ethidium bromide. Successful PCR amplification products resulted in a single DNA band (corresponding to ~560 bp). The PCR products were purified using a Biolight PCR Purification Kit (Shanghai Biolight Technology Co. Ltd., China) according to the manufacturer’s instructions.
2.5. Sequencing and analysis of rDNA-ITS regions
The purified PCR products were submitted to the Hangzhou Genomics Institute for sequencing in both directions. Sequence files were assembled and edited, and consensus sequences were constructed using DNAMAN 4.0 (Lynnon bioSoft). The ITS sequences were submitted to the GenBank database. Sequences were deposited in GenBank (accession numbers given in Table 1) and compared against those sequences already found in the databases using the BLAST search. The ITS sequences for reference isolates were downloaded from the GenBank database (Table 1), along with that of Neurospora crassa (M13906) as an outgroup. Sequences were aligned using CLUSTAL X (Thompson et al., 1997). Phylogenetic analyses were performed using test version 4.0b10 (PPC) of PAUP (Swofford, 2002). Phylogenetic trees were inferred from the ITS sequence data set using parsimony analysis and all characters were weighted equally and gaps were treated as missing data. Clade stability was assessed using a bootstrap analysis with 1 000 replicates, each with 10 replicates of random stepwise addition of taxa.
Table 1.
Isolates of Colletotrichum spp. from Zhejiang Province and Shanghai City, China used in this study and the reference isolates used for ITS ribosomal DNA sequence comparisons
| Isolate/strain | Species | Host plant | Geographical origin | GenBank accession No. | Reference |
| Isolates used | |||||
| WY01 | C. gloeosporioides | Toyonoka/leaf* | Jiande City, Zhejiang Province | FJ608625 | In this study |
| WY02 | C. gloeosporioides | Toyonoka/petiole | Jiande City, Zhejiang Province | FJ608626 | In this study |
| WY03 | C. gloeosporioides | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608627 | In this study |
| WY04 | C. gloeosporioides | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608628 | In this study |
| WY05 | C. gloeosporioides | Toyonoka/leaf | Zhaotun, Shanghai City | FJ608629 | In this study |
| WY06 | C. gloeosporioides | Toyonoka/crown | Zhaotun, Shanghai City | FJ608630 | In this study |
| WY07 | C. gloeosporioides | Hongjia/leaf | Hangzhou City, Zhejiang Province | FJ608631 | In this study |
| WY08 | C. gloeosporioides | Toyonoka/leaf | Hangzhou City, Zhejiang Province | FJ608632 | In this study |
| WY09 | C. gloeosporioides | Toyonoka/crown | Jiande City, Zhejiang Province | FJ608633 | In this study |
| WY10 | C. gloeosporioides | Toyonoka/petiole | Hangzhou City, Zhejiang Province | FJ608634 | In this study |
| WY11 | C. fragariae | Toyonoka/crown | Jiande City, Zhejiang Province | FJ608635 | In this study |
| WY12 | C. fragariae | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608636 | In this study |
| WY13 | C. fragariae | Toyonoka/crown | Zhaotun, Shanghai City | FJ608637 | In this study |
| WY14 | C. fragariae | Toyonoka/leaf | Zhaotun, Shanghai City | FJ608638 | In this study |
| WY15 | C. fragariae | Toyonoka/crown | Zhaotun, Shanghai City | FJ608639 | In this study |
| WY16 | C. fragariae | Tochiotome/crown | Hangzhou City, Zhejiang Province | FJ608640 | In this study |
| WY17 | C. fragariae | Toyonoka/leaf | Hangzhou City, Zhejiang Province | FJ608641 | In this study |
| WY18 | C. fragariae | Akihime/petiole | Hangzhou City, Zhejiang Province | FJ608642 | In this study |
| WY19 | C. fragariae | Toyonoka/leaf | Hangzhou City, Zhejiang Province | FJ608643 | In this study |
| WY20 | C. fragariae | Toyonoka/crown | Jiande City, Zhejiang Province | FJ608644 | In this study |
| WY21 | C. acutatum | Toyonoka/leaf | Zhaotun, Shanghai City | FJ608645 | In this study |
| WY22 | C. acutatum | Toyonoka/fruit | Zhaotun, Shanghai City | FJ608646 | In this study |
| WY23 | C. acutatum | Toyonoka/leaf | Zhaotun, Shanghai City | FJ608647 | In this study |
| WY24 | C. acutatum | Toyonoka/fruit | Zhaotun, Shanghai City | FJ608648 | In this study |
| WY25 | C. acutatum | Toyonoka/fruit | Jiande City, Zhejiang Province | FJ608649 | In this study |
| WY26 | C. acutatum | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608650 | In this study |
| WY27 | C. acutatum | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608651 | In this study |
| WY28 | C. acutatum | Toyonoka/fruit | Jiande City, Zhejiang Province | FJ608652 | In this study |
| WY29 | C. acutatum | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608653 | In this study |
| WY30 | C. acutatum | Toyonoka/leaf | Jiande City, Zhejiang Province | FJ608654 | In this study |
| WY31 | C. acutatum | Toyonoka/petiole | Jiande City, Zhejiang Province | FJ608655 | In this study |
| Reference isolates | |||||
| IMI 345047 | C. fragariae | Strawberry | USA | AJ536223 | Martinez-Culebras et al., 2003 |
| IMI 348167 | C. fragariae | Strawberry | USA | AJ536222 | Martinez-Culebras et al., 2003 |
| CG 231 | C. gloeosporioides | Strawberry | USA | AF272780 | Freeman et al., 2001 |
| IMI 324983 | Glomerella cingulata | Strawberry | USA | AJ536226 | Martinez-Culebras et al., 2003 |
| IMI 347765 | G. tucumanensis | Saccharum sp. | Bangladesh | AJ536231 | Martinez-Culebras et al., 2003 |
| IMI 345051 | G. cingulata | Strawberry | Canada | AJ536224 | Martinez-Culebras et al., 2003 |
| IMI 345027 | G. acutata | Strawberry | France | AJ536199 | Martinez-Culebras et al., 2003 |
| IMI 348160 | G. acutata | Strawberry | USA | AJ536200 | Martinez-Culebras et al., 2003 |
| IMI 360086 | G. acutata | Strawberry | Japan | AJ536205 | Martinez-Culebras et al., 2003 |
| IMI 345575 | G. acutata | Strawberry | New Zealand | AJ536215 | Martinez-Culebras et al., 2003 |
| MAFF 511343 | G. graminicola | unknown | Japan | AB057436 | Moriwaki et al., 2002 |
| CBS 151.28 | G. lindemuthiana | Phaseolus vulgaris | unknown | AJ301946 | Nirenberg et al., 2002 |
| BBA 70709 | C. trifolii | Trifolium sp. | USA | AJ301941 | Nirenberg et al., 2002 |
| UQ 343 | C. destructivum | Unknown | Australia | AF451908 | Unpublished |
| STE-U 5304 | C. capsici | Arachis hypogaea | Tanzania | AY376526 | Lubbe et al., 2004 |
| STE-U 5301 | C. coccodes | Lycopersicon esculentum | Zimbabwe | AY376528 | Lubbe et al., 2004 |
| STE-U 5299 | C. dematium | Peperomia sp. | Unknown | AY376531 | Lubbe et al., 2004 |
| BBA 70535 | C. fuscum | Digitalis lanata | unknown | AJ301938 | Nirenberg et al., 2002 |
| STE-U 5296 | C. orbiculare | Xanthium spinosum | Argentina | AY376541 | Lubbe et al., 2004 |
| STE-U 5294 | G. truncata | Arachis hypogaea | Gambia | AY376543 | Lubbe et al., 2004 |
Cultivar/Organ from which fungus isolated
2.6. PCR with C. acutatum-specific primers
PCR primers for detection of the pathogen included the ITS4 primer (5′-TCCTCCGCTTATTGATATGC-3′) coupled with the specific primer for C. acutatum (CaInt2) (5′-GGGGAAGCCTCTCGCGG-3′) (Sreenivasaprasad et al., 1996; White et al., 1990). The genomic DNA of Trichoderma virens was used as a negative control. C. acutatum-specific PCR reactions were performed in a total volume of 50 μl, containing 50 ng of genomic DNA, 50 mmol/L KCl, 10 mmol/L Tris-HCl, 100 μmol/L of each dNTP, 1.5 mmol/L MgCl2, 1 U of Taq DNA polymerase (TaKaRa Corp., Japan), and 0.5 μmol/L of each primer. The reaction mixtures were incubated in a PTC-150 MiniCycler (MJ RESEARCH Corp., America) for 35 cycles consisting of 1 min at 95 °C, 30 s at 54 °C, and 1 min at 72 °C. Amplification products were separated in 1% (w/v) agarose gels, stained with the nucleic acid stain GoldView™ (Shanghai SBC Genetch Co. Ltd., China), and viewed under UV light for detection of amplification products.
2.7. PCR reaction and DNA digestion
The 5.8S-ITS regions of representative isolates of C. gloeosporioides and C. fragariae, identified by morphological characteristics, were amplified by PCR using universal primers ITS1 (5′-GCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Martinez-Culebras et al., 2000). The genomic DNA of Trichoderma virens was used as a negative control. PCR reactions were performed in a total volume of 50 μl, containing 150–200 ng of DNA, 50 mmol/L KCl, 10 mmol/L Tris-HCl, 200 μmol/L of each dNTP, 1 μmol/L of each primer, 1.5 mmol/L MgCl2, and 2.5 U of Taq DNA polymerase (Sangon Co., Shanghai, China). The reaction mixtures were incubated in a PTC-150 MiniCycler (MJ RESEARCH Corp., America). Following an initial denaturation at 95 °C for 4 min, the DNA templates were amplified for 35 cycles. Each cycle consisted of a denaturation step at 95 °C for 1 min, an annealing step at 56 °C for 1 min, and an extension step at 72 °C for 1 min. A final extension step (72 °C for 10 min) was included at the end. Amplification products were separated on 1% (w/v) agarose gels in Tris-acetate-EDTA (TAE) buffer.
To specifically identify strawberry isolates of C. gloeosporioides, PCR products from the ITS1/ITS4 primer pair were digested with the restriction enzyme MvnI (supplied under the name Bsh1236I by Fermentas, MBI, Republic of Lithuania) at 37 °C for 16 h (Martinez-Culebras et al., 2003). PCR products for species-specific identification and restriction fragments from the PCR fragment primed with ITS1 and ITS4 were separated on 1% and 3% (w/v) agarose gels, respectively, with 0.5× Tris-borate-EDTA (TBE) buffer. After electrophoresis, gels were stained with ethidium bromide (0.5 μg/ml), and the DNA bands were visualized with UV light. DNA sizes were estimated by comparison with a DNA length standard (100–3 000 bp molecular marker, Bio Basic Inc., Canada), as described by Martinez-Culebras et al. (2003).
3. Results
3.1. Morphological comparisons
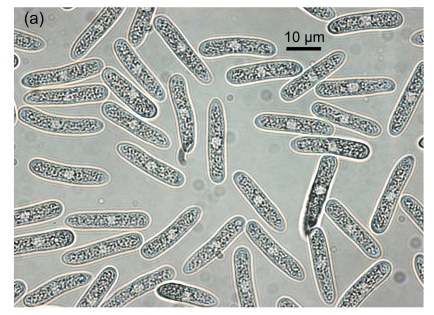
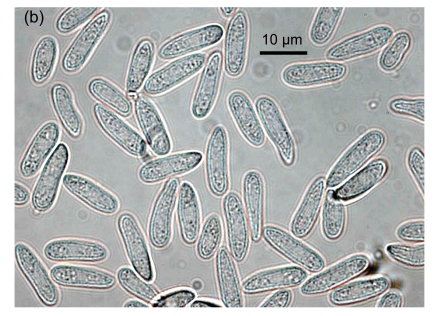
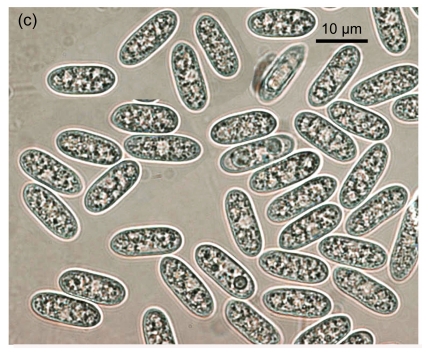
Isolates of Colletotrichum spp. obtained from fruit, vegetative parts, and root crowns from different cultivars showing typical anthracnose symptoms, were selected for morphological identification. Based on taxonomic criteria of morphological characteristics (colony characters, conidial and setal morphology and production of a teleomorph) (Gunnell and Gubler, 1992), 10 C. gloeosporioides, 10 C. fragariae and 11 C. acutatum isolates were recognized among 31 isolates (Table 1). On SLA, most of the conidia of C. fragariae isolates were narrowly obovate, straight or occasionally slightly curved, [(13.0–) 14.5–16.5 (–19.0)] μm×[(3.5–) 4.2–4.8 (–5.5)] μm (mean=16.0 μm×4.5 μm) (Fig. 1a). Conidia of C. acutatum isolates were elliptic to fusiform, [(11.0–) 11.2–15.5 (–16.0)] μm×[(3.8–) 4.1–4.7 (–5.0)] μm (mean=13.5 μm×4.4 μm) (Fig. 1b). The conidia of C. gloeosporioides isolates were oblong with obtuse ends, straight (Fig. 1c), and were shorter and broader than those of C. fragariae and C. acutatum, [(11.8–) 12.5–14.8 (–16)] μm×[(4.6–) 5.0–6.0 (–6.8)] μm (mean=13.5 μm×5.5 μm). The shapes of the conidia of C. fragariae, C. gloeosporioides and C. acutatum agreed with previous descriptions (Gunnell and Gubler, 1992), but the average conidial size of each of the three species was slightly different from the range described by Gunnell and Gubler (1992), namely 18 μm×4 μm, 15.5 μm×3.7 μm, and 15 μm×4.3 μm for C. fragariae, C. acutatum, and C. gloeosporioides, respectively.
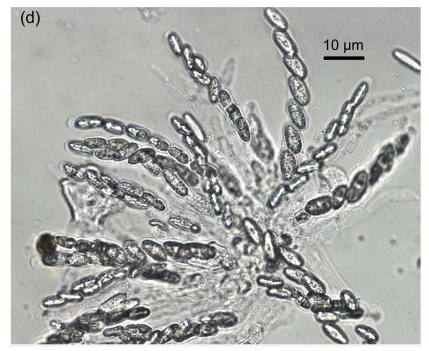
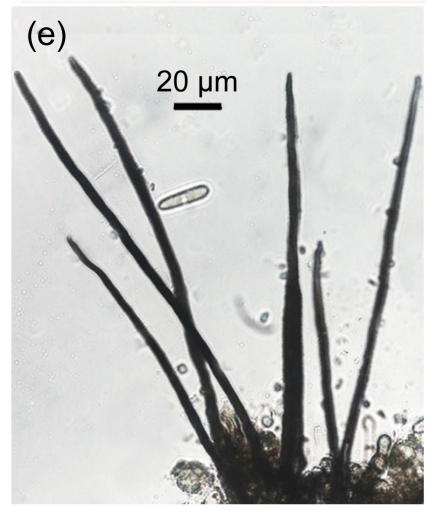
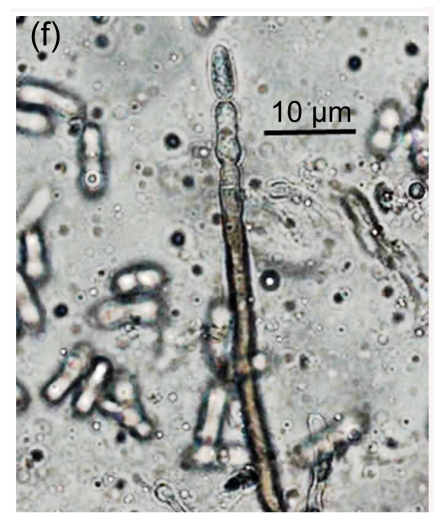
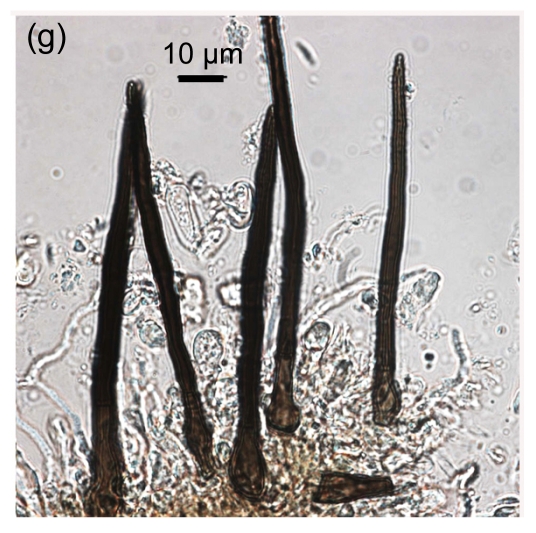
Fig. 1.

Conidial morphology of Colletotrichum species isolates from strawberry. (a) Obovate conidia of C. fragariae; (b) Elliptic to fusiform conidia of C. acutatum; (c) Cylindrical conidia of C. gloeosporioides; (d) Asci and ascospores of C. gloeosporioides; (e) & (f) Setae of C. fragariae; (f) A seta producing a conidium from its apical cells; (g) Setae of C. gloeosporioides; (h) Seta of C. acutatum
Mature setae of C. fragariae were dark brown, uniform in width except for the apical cells which functioned as phialides, and produced conidia in individual isolates (Figs. 1e and 1f). Setae of C. gloeosporioides were dark brown, gradually tapered along their entire length to their apices, straight, produced singly, and did not produce conidia (Fig. 1g). Setae of C. acutatum were brown to dark brown, tapered, generally aseptate (Fig. 1h), did not produce conidia, and were shorter than those of C. fragariae and C. gloeosporioides. The isolates of C. gloeosporioides readily produced perithecia containing asci and ascospores (Fig. 1d), but none of the isolates of C. fragariae or C. acutatum produced perithecia.
On PDA, the colony color of C. fragariae isolates ranged from beige to dark grey. Colonies of C. gloeosporioides isolates showed a dense, white mycelial growth turning to a dark olive-grey color. The colony color of C. acutatum isolates was white for 4–5 d but later became grey-brown.
3.2. Sequence analysis of the ribosomal DNA spacer sequence
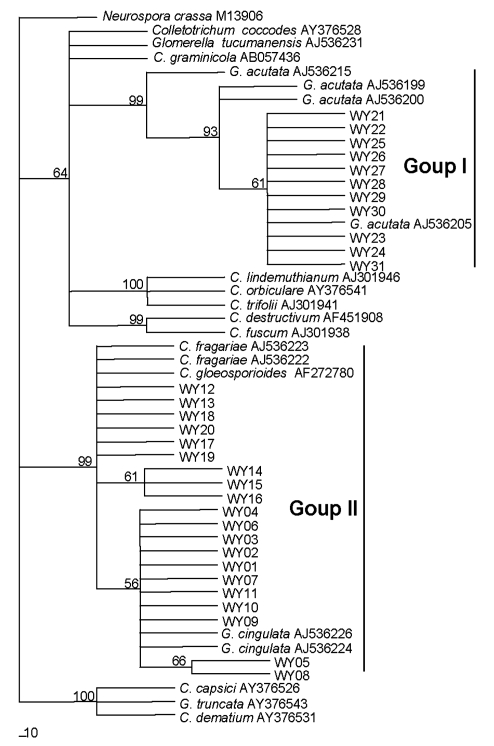
Sequence alignment showed that among 11 isolates (WY21–WY31), all sequences were identical to that of a strawberry isolate of C. acutatum (IMI 360086) from Japan. Among the other isolates (WY01–WY20), all sequences were very similar.
Reference sequences were selected cautiously from GenBank, with only those from voucher specimens being selected. The final ITS/5.8S dataset included 52 sequences with 530 characters after alignment. Phylogenetic analysis showed that the 31 isolates in this study were clustered into two groups, group I and group II (Fig. 2). In group I, 11 isolates, identified as C. acutatum based on morphological characteristics, were clustered together with 4 reference isolates of C. acutatum (teleomorph G. acutata) from strawberry. The isolate IMI 345575 (New Zealand) formed a single subgroup and did not group with any others. While 11 isolates produced a clade with the reference sequence of C. acutatum IMI 360086 from Japan, they were clustered with the isolates IMI 348160 (USA) and IMI 345027 (France) to form another subgroup with 93% bootstrap support, as described in Table 1 of Sreenivasaprasad and Talhinhas (2005). In group II, 20 isolates (WY01–WY20), classified as C. fragariae and C. gloeosporioides based on morphology, were in a poorly resolved group that included the reference isolates of C. fragariae and C. gloeosporioides (teleomorph G. cingulata) from strawberry (Fig. 2).
Fig. 2.
Phylogram generated from parsimony analysis based on complete sequence of ITS1-5.8S-ITS2 ribosomal RNA genes of 52 sequences with Neurospora crassa as outgroup
Data were analyzed with random addition sequence, unweighted parsimony and treating gaps as missing data. Bootstrap values ≥50% are shown above or below branches. The numbers near each branch represent percentages out of 100 bootstrap replications
However, by analyzing 5.8S-ITS sequences, an MvnI restriction site was found to be present within the ITS1 region in only 10 isolates (WY01–WY10) and no MvnI restriction sites were found in the other isolates. Previous studies have shown an MvnI restriction site present within the ITS1 region of the 5.8S-ITS sequences of only C. gloeosporioides isolates but not in other isolates of Colletotrichum spp. from strawberry plants (Martinez-Culebras et al., 2000; 2003). So in group II, 10 isolates (WY01–WY10) were related to C. gloeosporioides and the other isolates were attributed to C. fragariae, consistent with morphological identification (Table 1).
The ITS1-5.8S-ITS2 rDNA sequences of the 10 C. gloeosporioides isolates fell into two groups: the first group comprised sequences from eight isolates (WY01, WY02, WY03, WY04, WY07, WY09, WY10 and WY06) and the second group comprised sequences from the remaining two (WY05 and WY08). Sequences within each group were identical but the two groups differed by three base pairs. Sequences of the C. fragariae isolates fell into three groups comprising four (WY18, WY20, WY12 and WY13), three (WY14, WY15 and WY16), and three (WY19, WY17 and WY11) isolates, respectively. Again, sequences within each group were identical but the three groups differed by three base pairs. Analysis of the rDNA ITS sequences revealed that the MvnI restriction site in C. gloeosporioides isolates was conservative and a useful tool for distinguishing C. gloeosporioides from C. fragariae from strawberry plants. It also showed the genetic diversity of population structures in C. gloeosporioides and C. fragariae.
Based on 5.8S-ITS sequences and phylogenetic analysis, the 31 isolates could be classified clearly into three species. Eleven isolates (WY21–WY31) were identified as C. acutatum (all identical), 10 (WY01–WY10) as C. gloeosporioides (8 clustering in a group that is 3-bp different from the remaining 2) and 10 (WY11–WY20) as C. fragariae (4 clustering in a group that is 3-bp different from the remaining 6 that are in two groups of 3), consistent with the results of morphological identification (Table 1). Based on host origin, 7 isolates of C. acutatum, 8 of C. gloeosporioides and 7 of C. fragariae were isolated from the Zhejiang area, and 4, 2 and 3, respectively, from the Shanghai region. This confirms that the species of Colletotrichum causing anthracnose disease of strawberry in Zhejiang and Shanghai are the same.
3.3. Species-specific PCR and enzyme digestion
Representative isolates, WY01, WY06 and WY08 for C. gloeosporioides, WY12, WY19 and WY14 for C. fragariae, and WY27, WY29 and WY31 for C. acutatum, were selected for species-specific PCR and enzyme digestion.
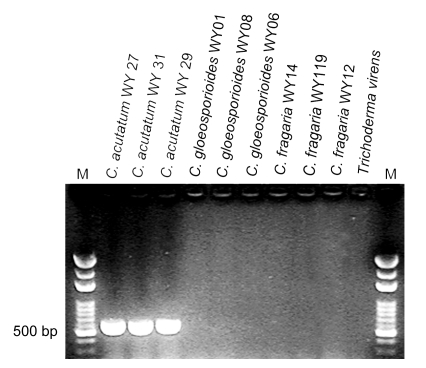
The nine representative isolates were detected by using the C. acutatum-specific primer CaInt2 in conjunction with the ITS4 primer. About 490-bp fragments were amplified from the genomic DNA of the C. acutatum isolates but no amplification products were produced from the isolates representative of C. gloeosporioides or C. fragariae, along with the negative control (Trichoderma virens) (Fig. 3). This species-specific PCR result further confirmed the species identity of C. acutatum, consistent with phylogenetic and morphological identification.
Fig. 3.
PCR amplification of a specific fragment from Colletotrichum species using the C. acutatum specific primer CaInt2 in conjunction with the primer ITS4
Lanes 1–3 correspond to representative C. acutatum isolates; Lanes 4–6 are the representative C. gloeosporioides isolates; Lanes 7–9 correspond to representative C. fragariae isolates; Lane 10 is a negative control (Trichoderma virens genomic DNA); Lane M corresponds to the 100–3 000 bp molecular weight marker (Bio Basic Inc., Canada)
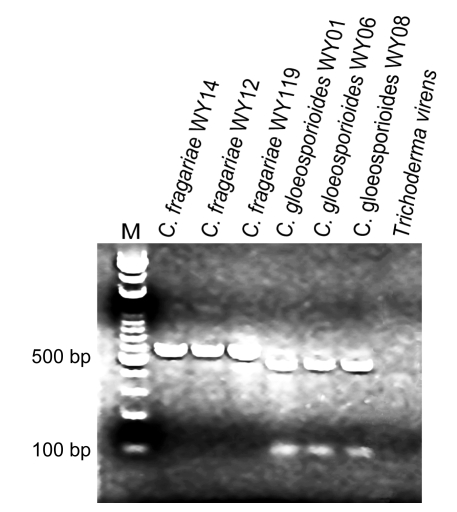
The isolates representative of C. fragariae and C. gloeosporioides were analyzed using the endonuclease MvnI. The restriction endonuclease MvnI cut all C. gloeosporoides PCR products amplified by PCR using the universal primers ITS1 and ITS4 to produce two fragments of about 480 and 100 bp. The species-specific restriction pattern could be obtained from the C. gloeosporioides isolates but this endonuclease did not cut any of the C. fragariae isolates (Fig. 4). This result confirms the established method for species identification using MvnI digestion data (Martinez-Culebras et al., 2000; 2003).
Fig. 4.
Ribosomal DNA restriction patterns shown by Colletotrichum species after amplification with universal primers ITS1 and ITS4 followed by digestion with the restriction endonuclease MvnI
Lanes 1–3 correspond to representative C. fragariae isolates; Lanes 4–6 correspond to representative C. gloeosporioides isolates; Lane 7 is a positive control (Trichoderma virens genomic DNA); Lane M corresponds to the 100–3 000 bp molecular weight marker (Bio Basic Inc., Canada)
4. Discussion
Most strawberry cultivars grown in Zhejiang Province and Shanghai City were introduced from Japan and the cv. Toyonoka is presently the principal cultivar. Almost all of the isolates of Colletotrichum spp. responsible for the anthracnose disease originated from this cultivar. In this study, we identified the Colletotrichum spp. causing strawberry anthracnose, and aimed to determine whether the same or different species occur in strawberry nurseries and production fields in Zhejiang and Shanghai.
Traditionally, three species from strawberry were considered to be differentiated by morphological characters (Gunnell and Gubler, 1992). However, morphological criteria can be unreliable and non-predictive, because of highly variable morphological characteristics (Sreenivasaprasad et al., 1996; Sutton, 1992; Johnston and Jones, 1997). So Ye et al. (1997) possibly failed to distinguish C. gloeosporioides from C. fragariae from strawberry plants from Shanghai.
Molecular genetic studies have provided useful data for clarifying the systematics of the genus Colletotrichum (Martinez-Culebras et al., 2003). However, our studies showed that the divergence of 5.8S-ITS region sequences among isolates of C. fragariae and C. gloeosporioides was too low to separate them confidently as different species, confirming the observations of Sherriff et al. (1995), Sreenivasaprasad et al. (1996) and Martinez-Culebras et al. (2003). Similarly, Ren et al. (2008) found that the ITS sequences of 17 isolates from strawberry were 99% identical, but they all were mistaken for C. gloeosporioides. Although the similarity between the nucleotide sequences of C. gloeosporioides and C. fragariae prevented the development of specific primers for these species based on the 5.8S-ITS region, Martinez-Culebras et al. (2000) found that there was an MvnI specific site among ITS1 region of isolates of only C. gloeosporioides and not C. fragariae. This trait was confirmed subsequently by different original sequence data and could be used for differentiating C. gloeosporioides from C. fragariae (Martinez-Culebras et al., 2003). Martinez-Culebras et al. (2003) pointed out that this conclusion still needed to be confirmed by studying a higher number of isolates of C. fragariae and C. gloeosporioides from strawberry and also C. gloeosporioides from different hosts (Martinez-Culebras et al., 2003). Our results provide new evidence that the MvnI site within the ITS1 region from isolates of C. gloeosporioides from strawberry is conservative.
In C. acutatum, phylogenetic analysis based on 5.8S-ITS region sequences indicated that different original isolates of pathogens of strawberry form a polyphyletic taxon, consistent with other observations (Martinez-Culebras et al., 2003; Freeman et al., 2000; 2001). The sequences of our 11 isolates of C. acutatum from Zhejiang and Shanghai, however, were identical to that of a strawberry isolate of C. acutatum (IMI 360086) from Japan. Although currently there are over 600 rDNA-ITS sequences from C. acutatum isolates available in public databases, a BLAST search showed that our sequences were identical to those of AL-9 (GenBank accession No. AB219025), AL-5 (GenBank accession No. AB219024) and IMI 360086 found only on strawberry in Japan. This suggests that this species has been introduced into Zhejiang and Shanghai from Japan through infected propagation material and has rapidly become established locally. However, analysis of sequences showed that there were two sequence types in C. gloeosporioides and three in C. fragariae. These may reflect the genetic diversity of population structures within them. Within C. gloeosporioides, two sequence types were distributed in Zhejiang and Shanghai but based on the frequency of isolation, and the sequence types of the isolates WY01, WY02, WY03, WY05, WY06, WY07, WY09 and WY10 might be the predominant forms. Within C. fragariae, there were three sequence types of which two were distributed in Zhejiang and Shanghai and one, from isolates WY19, WY17 and WY11, was found only in Zhejiang (Table 1).
Based on isolation data (Table 1), C. acutatum can be isolated from fruits, leaves and petioles, C. gloeosporioides from leaves, petioles and root crowns, and C. fragariae from root crowns, leaves and petioles. Differences in pathogenicity among them, however, still need to be evaluated by pathogenicity tests. Although pathogenicity tests had been performed and typical symptoms could be seen, control transplant seedlings also could produce similar symptoms. Apparently, Colletotrichum spp. may be present in the form of latent infections in strawberry. The presence of latent infections of Colletotrichum spp. in strawberry was detected by a freeze treatment method and Colletotrichum spp. could be found on up to 50% of leaves and 10% of petioles in transplant seedlings (Mertely and Legard, 2004). For this reason, currently we still have not obtained reliable results from pathogenicity tests. However, this may be resolved in future studies by using pathogen-free strawberry plants obtained by propagating plantlets from meristem culture.
Based on morphological characters, phylogenetic analysis, species-specific PCR and enzyme digestion, three species, C. acutatum, C. gloeosporioides and C. fragariae, are considered the cause of anthracnose in strawberry-growing areas of Zhejiang and Shanghai. Although molecular studies are helpful for resolving relationships within the genus Colletotrichum, studies to date can be regarded as only preliminary (Photita et al., 2005). Many questions remain regarding the interspecies relationships within the genus, such as between C. gloeosporioides and C. fragariae. Sequence analysis from ITS regions has enabled some progress to be made towards a better understanding of the taxonomy of Colletotrichum, but the analysis of a larger collection of C. gloeosporioides and C. fragariae isolates, preferably based on a multilocus comparison (e.g., of ITS, actin, beta-tubulin, calmodulin, glutamine synthetase), is required before the two taxa can be differentiated with confidence.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30571208) and the Key Scientific and Technological Project of Hangzhou City (No. 200432239), China
References
- 1.Buddie A, Martínez-Culebras P, Bridge P, García M, Querol A, Cannon P, Monte E. Molecular characterization of Colletotrichum strains derived from strawberry. Mycological Research. 1999;103(4):385–394. doi: 10.1017/S0953756298007254. [DOI] [Google Scholar]
- 2.Cooke DEL, Duncan JM. Phylogenetic analysis of Phytophthora species based on the ITS1 and ITS2 sequences of ribosomal DNA. Mycological Research. 1997;101(6):667–677. doi: 10.1017/S0953756296003218. [DOI] [Google Scholar]
- 3.Dai FM, Ren XJ, Lu JP. First report of anthracnose fruit rot of strawberry caused by Colletotrichum acutatum in China. Plant Disease. 2006;90(11):1460. doi: 10.1094/PD-90-1460A. [DOI] [PubMed] [Google Scholar]
- 4.Debode J, Hemelrijck W, Baeyen S, Creemers P, Heungens K, Maes M. Quantitative detection and monitoring of Colletotrichum acutatum in strawberry leaves using real-time PCR. Plant Pathology. 2009;58(3):504–514. doi: 10.1111/j.1365-3059.2008.01987.x. [DOI] [Google Scholar]
- 5.Denoyes-Rothan B, Guérin G, Délye C, Smith B, Minz D, Maymon M. Genetic diversity and pathogenic variability among isolates of Colletotrichum species from strawberry. Phytopathology. 2003;93(2):219–228. doi: 10.1094/PHYTO.2003.93.2.219. [DOI] [PubMed] [Google Scholar]
- 6.Freeman S, Minz D, Jurkevitch E, Maymon M, Shabi E. Molecular analysis of Colletotrichum species from almond and other fruits. Phytopathology. 2000;90(6):608–614. doi: 10.1094/PHYTO.2000.90.6.608. [DOI] [PubMed] [Google Scholar]
- 7.Freeman S, Minz D, Maymon M, Zveibil A. Genetic diversity within Colletotrichum acutatum sensu Simmonds. Phytopathology. 2001;91(6):586–592. doi: 10.1094/PHYTO.2001.91.6.586. [DOI] [PubMed] [Google Scholar]
- 8.Gunnell PS, Gubler WD. Taxonomy and morphology of Colletotrichum species pathogenic to strawberry. Mycologia. 1992;84(2):157–165. doi: 10.2307/3760246. [DOI] [Google Scholar]
- 9.Johnston RP, Jones D. Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia. 1997;89(3):420–430. doi: 10.2307/3761036. [DOI] [Google Scholar]
- 10.Lubbe CM, Denman S, Cannon PF, Groenewald JZ, Lamprecht SC, Crous PW. Characterization of Colletotrichum species associated with diseases of Proteaceae. Mycologia. 2004;96(6):1268–1279. doi: 10.2307/3762144. [DOI] [PubMed] [Google Scholar]
- 11.MacKenzie SJ, Mertely JC, Seijo TE, Peres NA. Colletotrichum fragariae is a pathogen on hosts other than strawberry. Plant Disease. 2008;92(10):1432–1438. doi: 10.1094/PDIS-92-10-1432. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Culebras PV, Barrio E, García MD, Querol A. Identification of Colletotrichum species responsible for anthracnose of strawberry based on the internal transcribed spacers of the ribosomal region. FEMS Microbiology Letter. 2000;189(1):97–101. doi: 10.1016/S0378-1097(00)00260-3. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Culebras PV, Querol A, Suarez Fernandez MB, Garcia Lopez MD, Barrio E. Phylogenetic relationships among Colletotrichum pathogens of strawberry and design of PCR primers for their identification. Journal of Phytopathology. 2003;151(3):135–143. doi: 10.1046/j.1439-0434.2003.00694.x. [DOI] [Google Scholar]
- 14.Mertely JC, Legard DE. Detection, isolation, and pathogenicity of Colletotrichum spp. from strawberry petioles. Plant Disease. 2004;88(4):407–412. doi: 10.1094/PDIS.2004.88.4.407. [DOI] [PubMed] [Google Scholar]
- 15.Moriwaki J, Tsukiboshi T, Sato T. Grouping of Colletotrichum species in Japan based on rDNA sequences. Journal of General Plant Pathology. 2002;68(4):307–320. doi: 10.1007/PL00013096. [DOI] [Google Scholar]
- 16.Nirenberg HI, Feiler U, Hagendorn G. Description of Colletotrichum lupini comb. nov. in modern terms. Mycologia. 2002;94(2):307–320. doi: 10.2307/3761809. [DOI] [PubMed] [Google Scholar]
- 17.Photita W, Taylor PWJ, Ford R, Lumyong P, McKenzie HC, Hyde KD. Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand. Fungal Diversity. 2005;18:117–133. [Google Scholar]
- 18.Ren XJ, Liang Y, Lu JP, Yang BR, Xu JY, Dai FM. Identification of Colletotrichum species from strawberry in Shanghai. Acta Phytopathologica Sinica. 2008;38(3):325–328. [Google Scholar]
- 19.Shao XH. New disease of strawberry: strawberry anthracnose. Acta Agriculturae Shanghai. 1992;8(2):86–87. [Google Scholar]
- 20.Sherriff C, Whelan MJ, Arnold GM, Bailey JA. rDNA sequence analysis confirms the distinction between Colletotrichum graminicola and C. sublineolum . Mycological Research. 1995;99(4):475–478. doi: 10.1016/S0953-7562(09)80649-7. [DOI] [Google Scholar]
- 21.Sreenivasaprasad S, Talhinhas P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Molecular Plant Pathology. 2005;6(4):361–378. doi: 10.1111/j.1364-3703.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 22.Sreenivasaprasad S, Mills PR, Meehan BM, Brown AE. Phylogeny and systematics of 18 Colletotrichum species based on ribosomal DNA spacer sequences. Genome. 1996;39(3):499–512. doi: 10.1139/g96-064. [DOI] [PubMed] [Google Scholar]
- 23.Sutton BC. The Genus Glomerella and Its Anamorph Colletotrichum. In: Bailey JA, Jeger MJ, editors. Colletotrichum, Biology, Pathology and Control. Oxford, UK: CAB international; 1992. pp. 1–26. [Google Scholar]
- 24.Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods) Sunderland, USA: Sinauer Associates; 2002. Version 4b10. [Google Scholar]
- 25.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White TJ, Bruns SL, Taylor JW. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR Protocols. A Guide to Methods and Applications. San Diego: Academic Press Inc.; 1990. pp. 315–322. [Google Scholar]
- 27.Ye ZW, Zheng HQ, Tong YM. First report on species of strawberry anthracnose pathogen in Shanghai suburb and resistance of some strawberry cultivars to anthracnose. Acta Agriculturae Shanghai. 1997;13(1):75–80. [Google Scholar]