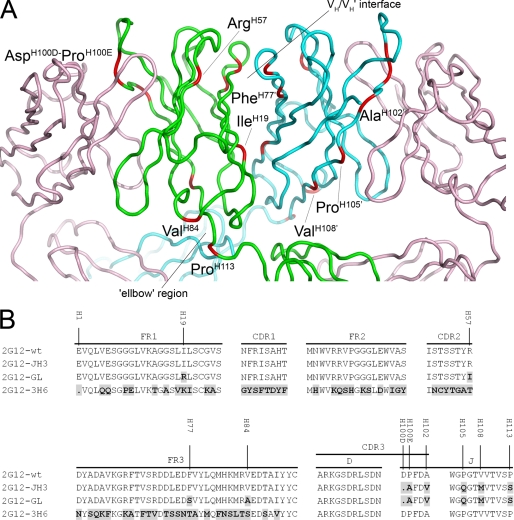
FIGURE 1.
2G12 Fab crystal structure A and amino acid alignment of the VDJ regions of 2G12 mutants compared with 2G12-wt B. A, the crystal structure of Fab 2G12 with the positions of the amino acid substitutions and deletions found in 2G12-GL shown in red and labeled. The light chains of the Fab dimer are shown in pink, with the heavy chains from Fab 1 and Fab 2 shown in green and blue, respectively. The VH/VH′ interface and the elbow region are also indicated (adapted from Protein Data Bank code 1op3) (6). B, differences in amino acid composition of the 2G12-JH3, 2G12-GL, and 2G12–3H6 mutants compared with 2G12-wt are shown in black with gray background. The amino acid numbering in A and B is based on the Kabat and Wu numbering system (19).