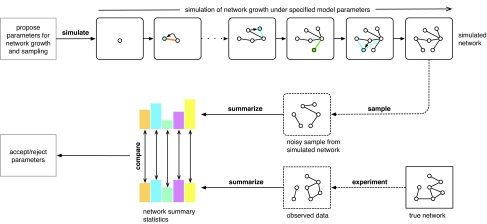
Figure 3. Repeated simulations under qualitative models of network growth can provide a starting point to explore plausible genome-wide modes of network evolution.
Networks are grown to the number of known proteins of a given organism, and binary interaction data sets are subsequently obtained under explicit assumptions of measurement error (Wiuf and Ratmann, 2009). These simulations are compared to the observed data in terms of summary statistics, such as those in Box 2: for methods that help in choosing summaries, we refer to (Ratmann et al., 2007; Joyce and Marjoram, 2008). ABC under model uncertainty (Ratmann et al., 2009) provides a Bayesian framework for these comparisons, and enables the inference of posterior distributions of the model parameters and summary errors. Crucially, the latter may provide information on model adequacy and the interpretability of the model parameters.