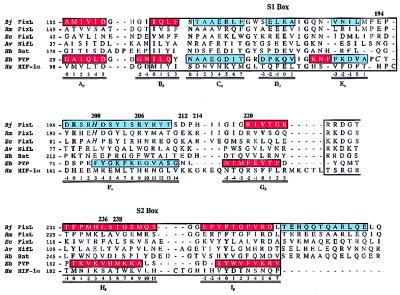
Figure 2.
Sequence alignment of selected PAS-domain proteins from Bacteria, Archaea, and Eukarya. The sequences correspond to the FixLs from B. japonicum (Bj FixL), Rhizobium meliloti (Rm FixL), and E. coli (Ec FixL), the NifL flavoprotein from Azotobacter vinelandii (Av NifL), the bacterio-opsin activator protein from Halobacterium halobium (Hh Bat), the PYP from Ectothiorhodospira halophila (Eh PYP), and the hypoxia-inducible factor-1α from humans (Hs HIF-1α) (4, 8, 9, 16, 38, 39). The S1 and S2 boxes (large boxes), conserved PAS-domain residues (bold type), sites of cofactor attachment (italics), α-helices (blue), and β-sheets (red) are shown. The secondary-structure assignments for met- and cyanomet-BjFixLH were determined by procheck and dssp (23, 40). The proposed naming scheme for secondary-structure regions is indicated below the alignment. The FixL sequences illustrate the conservation of residues specific to the regulatory mechanism. The other proteins were chosen to illustrate the ubiquity of the PAS fold in Bacteria, Archaea, and Eukarya, and the diversity of its functions and cofactors.