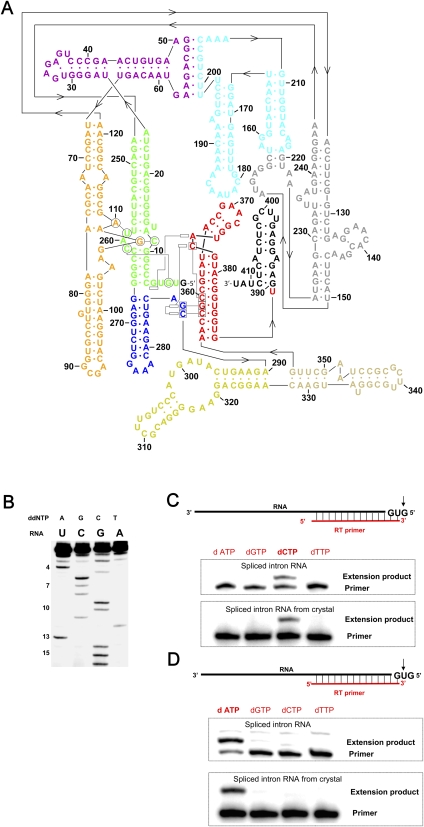
FIGURE 1.
Secondary structure and biochemical analysis of the spliced Oceanobacillus iheyensis intron. (A) Secondary structure of the O. iheyensis intron, shown in the revised representation (Toor et al. 2008a), which includes base-pairing and tertiary interaction designations derived from the refined structure presented here. The “classical view” of the secondary structure can be seen in Toor et al. (2008a), Figure 1A. The different domains and regions of the intron are indicated by color, in the same manner as previously published (Toor et al. 2008a): (green) domains I(i) and I(ii); (purple) domains IA and IB; (orange) domain IC; (gray) domain ID1; (cyan) domain ID2; (blue) domain DII; (yellow) domain DIII; (beige) domain DIV; (red) domain DV; and (black) domain DVI (not visualized in the crystal structure). (B) Reverse-transcriptase (RT) sequencing of the 5′-end of the spliced O. iheyensis intron. Nucleotide positions from the 5′-end are indicated by numbers on the left side of the gel. Identification of the (C) first and (D) second nucleotides from the 5′-end of the spliced O. iheyensis intron (top) before and (bottom) after crystallization. The top and bottom gels in panels C and D exhibit slight differences in relative electrophoretic mobility between the primer and the extension product because the two sets of gels were run for different time periods.