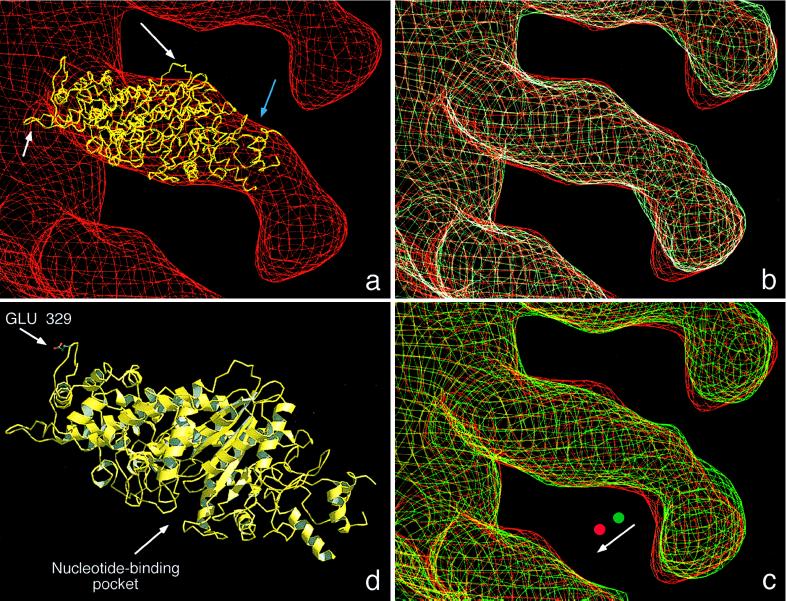
Figure 4.
(a) A molecular model of the catalytic domain of AMIC (yellow) was docked into the cryoelectron microscopy density map of the F-actin-AMIC (S329E) complex in rigor state (red). The model used for AMIC is the crystal structure of residues 80–783 of chicken smooth muscle myosin II (25). In the actin-proximal portion of the AMIC envelope, the model fits snugly, but toward the distal part of this domain (blue arrow), the volume is not fully occupied. The white arrows indicate two loops that protrude through the envelope, corresponding to sequences that are not conserved between the two myosins. Long arrow = D450 to Q456 of myosin II; short arrow = F366 to G379 of myosin II. (b) Superimposition of the molecular envelopes of the constitutively active mutant, S329E (red) and the constitutively inactive mutant, S329A (cyan), in rigor. (c) Superimposition of the constitutively active mutant, S329E, in its rigor (red) and ADP-bound (green) states. The shapes of the distal portion of the tail are essentially the same and suggest a small rigid-body rotation (see arrow) between the two states, about an axis near the midpoint of the myosin molecule. (d) Ribbon diagram of the model of the AMIC catalytic domain based on the crystal structure of chicken skeletal muscle myosin II (25). The positions of the regulatory residue at position 329 in the AMIC sequence (corresponding to position 411 in the muscle myosin) and of the nucleotide-binding pocket are marked. The model is in the same orientation from which AMIC is viewed in a–c. The diagram was prepared by using raster3d (26) and molscript (27).