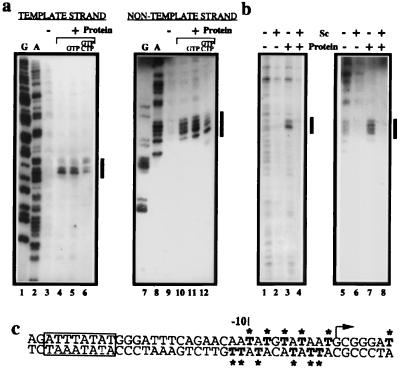
Figure 6.
Analysis of promoter opening by permanganate probing of the 16S promoter. (a) Open complex formation on negatively supercoiled 16S(σ = −0.053) promoter at 48°C. Lanes 1 and 7 and lanes 2 and 8 contain ddG and ddA sequence ladders, respectively. Lanes contain modification pattern in the absence of TBP, TFB, and RNAP (−, lanes 3 and 9), modification with TBP, TFB, RNAP, and no nucleoside triphosphates (+, lanes 4 and 10), modification with TBP, TFB, RNAP, and GTP (+, GTP; lanes 5 and 11), and modification with protein with GTP and CTP (+, G, CTP; lanes 6 and 12). Modification is detected on both template (lanes 1–6) and nontemplate strand (lanes 7–12). The region of modified thymidines is indicated by a vertical bar on the right. (b) No open complex is detectable on positively supercoiled templates at 48°C. Permanganate modification assays were performed on negatively supercoiled 16S (σ = −0.053, lanes 1, 3, 5, and 7) or positively supercoiled 16S (σ = +.047, lanes 2, 4, 6, and 8) DNA at 48°C in the presence (lanes 3, 4, 7, and 8) or absence (lanes 1, 2, 5, and 6) of TBP, TFB, and RNAP. (c) Summary of permanganate sensitivity data. The sequence of the 16S promoter is shown with the position of modified Ts indicated by bold type and ∗. The TATA element is boxed, and the site of transcription initiation is indicated by an arrow.