Abstract
Constraint based modeling is an approach for quantitative prediction of net reaction flux in genome scale biochemical networks. In vivo, the second law of thermodynamics requires that net macroscopic flux be forward, when the transformed reaction Gibbs energy is negative. We calculate the latter by using (i) group contribution estimates of metabolite species Gibbs energy, combined with (ii) experimentally measured equilibrium constants. In an application to a genome scale stoichiometric model of E. coli metabolism, iAF1260, we demonstrate that quantitative prediction of reaction directionality is increased in scope and accuracy by integration of both data sources, transformed appropriately to in vivo pH, temperature and ionic strength. Comparison of quantitative versus qualitative assignment of reaction directionality in iAF1260, assuming an accommodating reactant concentration range of 0.02 – 20 mM, revealed that quantitative assignment leads to a low false positive, but high false negative, prediction of effectively irreversible reactions. The latter is partly due to the uncertainty associated with group contribution estimates. We also uncovered evidence that the high intracellular concentration of glutamate in E. coli may be essential to direct otherwise thermodynamically unfavorable essential reactions, such as the leucine transaminase reaction, in an anabolic direction.
Keywords: systems biology, constraint-based modeling, thermodynamics
1 Introduction
Biological systems can be modeled at a large scale by taking an approach which balances computationally tractability with physically and biochemical realistic representation. Constraint-based modeling is a flexible and scalable approach for in silico phenotype prediction [1]. It relies on an accurate biochemical network reconstruction which is a biochemically, genetically and genomically structured representation of experimental biochemical & molecular biological literature [2]. In the case of metabolic networks, biochemical characterization of an enzyme establishes the substrate(s) and product(s) and genetic studies establish the gene-protein-reaction associations which tie a particular metabolic function in a model to a particular genomic location. A biochemical network reconstruction is then converted into a prototype computational model such that predictions may be compared with experimental data. In many cases, initial in silico tests suggest further refinements to the reconstruction underlying the prototype computational model. Iterative refinement of a constraint-based model, by comparison of prediction with experiment, supports its use for a priori in silico prediction of phenotypic capabilities for a posteriori in vivo experimental validation.
There have been many practical biological uses of constraint-based models, including study of bacterial evolution [3], analysis of network properties [4, 5, 6, 7], study of phenotypic behavior [8, 9], biological discovery [10, 11, 12], and metabolic engineering [13, 14, 15]. The growing scope of applications of genome scale metabolic reconstructions in metabolic engineering and other fields has recently been reviewed [16]. The predictive fidelity of a constraint-based model is dependent on the accuracy of the constraints used to eliminate physicochemically and biochemically infeasible network states. Generally, the resulting constraint equations define an under-determined feasible set of network states. Therefore, in unicellular organisms, a biological objective, such as maximization of growth rate, can be used to predict a single network state within this feasible set depending on the objective. The sensitivity of in silico predictions to the choice of objective function was treated in detail by Savinell & Palsson [17], and more recently by comparison of predictions with fluxomics data [18].
In this work, we focus on the assignment of reaction directionality in stoichiometric, metabolic models since it has a significant effect on the feasible set of functional states [19, 20, 21, 22]. There are two forms of thermodynamic constraints on reaction directionality. Local thermodynamic constraints apply on a reaction, by reaction basis. Essentially, a negative reaction Gibbs energy dictates a net forward reaction flux. This application of thermodynamics to the direction of biochemical reactions has a long history [23], with the first comprehensive treatment by Burton, Krebs and Kornberg [24]. Non-local thermodynamic constraints apply to sets of reactions [25] and arise due to the necessity to satisfy energy conservation, in addition to the second law of thermodynamics. Non-local thermodynamic constraints have been applied to small systems of biochemical reactions [26], but that approach “cannot be efficiently applied directly to genome-scale problems” [27] due to limitations imposed by computational complexity. Here, our focus is on local thermodynamic constraints for a system of reactions at genome scale. We summarize the theory underlying quantitative assignment of local reaction directionality at physiologically relevant conditions. Then, we apply this theory to a stoichiometric model of E. coli metabolism [28]. Our study relies on the extensive body of work on the thermodynamics of biochemical reactions by Alberty [29, 30] which we apply to a genome scale model for the first time. We complemented Alberty's approach with ongoing efforts by Henry et al. [19, 20] and Jankowski et al. [31] which seek to estimate the standard Gibbs energy of metabolite species based on a group contribution methodology.
2 Methods
2.1 Standard Gibbs energy of formation of metabolite species
There exist two complimentary quantitative methods for assigning reaction directionality based on different ways of calculating standard Gibbs energy of formation for metabolite species. The first involves back-calculation of standard Gibbs energy of formation using experimentally measured equilibrium constants [29]. In the absence of apparent equilibrium constants, standard Gibbs energy of formation for certain metabolite species cannot be back-calculated. In this case, a second complementary method involves estimation of standard Gibbs energy of formation of a metabolite species by summation of terms representing contributions to standard Gibbs energy of formation from different structural subgroups within that metabolite species [32, 31] (see Supplementary Material).
In constraint-based modeling of metabolic networks, it is typical to assume that all metabolite species concentrations are uniform throughout a single compartment enclosed by a lipid bilayer [33, 34]. This assumption is based on a local equilibrium hypothesis; that local variation in metabolite concentration is insignificant with respect to average compartment concentration [35]. In theory, a chemical reaction will perturb the spatial distribution of metabolite concentration, especially for certain metabolite intermediates at low concentration. However, unless the rate of such a reaction is diffusion limited, one can expect such perturbations to be insignificant on longer timescales. A significant number of publications on possible deviations of intracellular reaction thermodynamics from a local equilibrium hypothesis have been published and are reviewed in [36]. However, at present, due to the dearth of spatio-temporally resolved concentration data, one cannot be sure that any deviation from a local equilibrium hypothesis is actually significant in vivo. We assume the metabolite concentration is spatially invariant, and that, temperature and pressure are constant. Therefore one can define a single Gibbs energy of formation for each metabolite species which is valid throughout a single compartment.
Adjusting to in vivo ionic strength, temperature and pH
The standard Gibbs energy of formation of a metabolite species, , is an experimental measurement of the change in chemical potential which accompanies the synthesis of one mole of a metabolite species from its constituent elements in standard conditions. Standard conditions, denoted with the superscript, o, specify the synthesis of a one molar solution of a metabolite species from its constituent elements in their natural state, at temperature 298.15K, atmospheric pressure 1 bar, zero ionic strength and zero pH. Throughout this study, we assume atmospheric pressure and all thermodynamic potentials are expressed in kJ mol−1.
In our treatment of thermodynamic potentials, we assume that, with respect to metabolite species, the interior of a cell can be approximated by a buffer solution. This assumption neglects the fact that in general, there are effects on the equilibrium constants of in vivoreactions due to macromolecular crowding [37], confinement, adsorption and high fluid phase viscosity [38]. These effects are all a function of size and charge of the molecules involved. We confine ourselves to consideration of equilibrium constants for metabolic overall reactions between relatively small metabolites. Implicitly, with the exception of spontaneous reactions, all such reactions involve enzymes in elementary steps within the overall reactions. Enzymes are relatively large molecules but do not effect the equilibrium constant of an overall enzyme catalyzed reaction. For charged metabolites, there could be significant effects due to macromolecular adsorption and it is known that a higher intracellular fluid phase viscosity slows the rate of diffusion of metabolites [39]. However, in this study, we omit a treatment of these effects. The effect of macromolecular crowding and confinement is much more significant for macromolecules than metabolites and we do not consider these further. We do however consider the effects of in vivo pH, temperature and ionic strength.
In any quantitative system of relative measurements it is essential to have a standard reference point. However, any thermodynamic potential given for a ex vivo chemical standard reference condition must be adjusted to reflect in vivo conditions. The procedure for adjusting standard Gibbs energy of formation for each metabolite species, to in vivo ionic strength, temperature and pH, has been discussed theoretically and illustrated with computational implementation in textbooks by Alberty [29, 30]. We follow closely the same theoretical procedure, which is detailed in the Supplementary Material. There have been attempts to indirectly measure in vivo ionic strength, without actually measuring the concentrations of all charged metabolite species [40]. Such work would significantly refine our ability to model biochemical thermodynamics. However, at present, a pragmatic approach is to assume a physiological range of ionic strength between 0.05M and 0.35M [29], and then to adjust each metabolite species standard Gibbs energy of formation by assuming a particular ionic strength in this range.
We assume an ionic strength of 0.25 M then use the extended Debye-Hückel equation [41], to estimate the activity coefficient associated with each metabolite species, γj. The RT ln γi term is then absorbed into the standard Gibbs energy of formation at in vivo ionic strength. The advantage of this approach is that metabolomic data can be incorporated directly without adjusting it to represent ionic strength and charge dependent activities. The chemical standard reference temperature is typically room temperature, 298.15 K (25°C). However, generally for biochemical thermodynamics, reactions occur at body temperature, 310.15 K (37°C). It is possible to adjust metabolite species standard Gibbs energy of formation, within a narrow range of the chemical standard reference temperature, provided that the standard enthalpy of formation of a metabolite species is known, (See Supplementary Material).
Through a variety of homeostatic mechanisms [42], E. coli can grow over a wide range of external pH values, (5.5-9.0), yet maintains cytoplasmic pH between 7.6-7.8 [43, 44]. In contrast, the pH in the periplasmic space equals that of the external medium [44]. Metabolites, co-factors and ions make up approximately 3.5% of the dry weight of a typical E. coli cell [42]. While proteins account for approximately 55% of the dry weight. Cytoplasmic proteins represent a large buffering capacity, relative to the total pool of metabolites, co-factors and ions [45]. Effectively, the large buffering capacity of intracellular proteins maintains a constant concentration of cytoplasmic hydrogen ions. In a thermodynamic treatment of any system, knowledge of the values of a constant intensive variables dictates the appropriate thermodynamic potential to be used. This is concept applies regardless of whether the intensive variable is temperature or pH. As stressed by Alberty [29], the appropriate thermodynamic potential may be mathematically defined using a Legendre transform. However, knowledge of the latter is not essential to understand why it is appropriate to define a new thermodynamic potential, rather it is an elegant mathematical device for doing so. In the Supplementary Material, we show how to define a new thermodynamic potential for constant pH by adapting the classical thought experiment of a ‘bath’ that maintains an intensive variable constant.
Pseudo-isomer groups at specified pH
A reactant represents the properties of a set of related metabolite species. When pH is a known constant, a reactant may be treated as a pseudo-isomer group of metabolite species each of which is in a different state of hydrogen ion dissociation. e.g. at physiological ranges of pH, the reactant ATP consists of a series of different hydrogen ion dissociated forms. The concentration of the reactant ATP is given by the sum of the concentrations of the metabolite species in its pseudo-isomer group
Thus far, established practice in reconstruction of metabolic networks has been to approximate each reactant with a single predominant ionic form [28, 31]. Instead of this approximation, we follow the method of Alberty [29], and quantitatively amalgamate each property of a set of metabolite species into a property of the corresponding reactant. This retains the simplicity of qualitatively dealing with a single reactant but quantitatively representing the properties of a pseudo-isomer group.
Thermodynamically, at the timescale relevant for enzyme catalyzed reactions, the metabolite species which make up a single pseudo-isomer group may be considered to be in equilibrium with each other [46]. The hydrogen ion transfer rate constants range between 109 – 1011 s−1 M−1 as compared to the fastest enzyme turnover rates of ~ 107 s−1 M−1 [47]. Therefore, when modeling enzyme catalyzed reactions, it is reasonable to assume that the different metabolite species of a reactant are at equilibrium [29]. Thermodynamic state variables [41] for each metabolite species, such as standard Gibbs energy of formation, , must be individually adjusted from chemical standard conditions, to in vivo pH, ionic strength, and temperature. Thereafter, assuming equilibrium between metabolite species in a pseudo-isomer group, the adjusted state variables for each metabolite species in a pseudo-isomer group are then combined, and represented by a reactant state variable. The reactant state variable we require is the standard transformed Gibbs energy of a reactant, . Note that we distinguish a reactant, with index i, from a metabolite species with index j. Note also, the slight change in superscript, from o to 0, which is used to remind the reader that the latter is now a function of ionic strength (See supplementary material A.3).
The standard transformed Gibbs energy of formation of a reactant can be calculated by assuming that each metabolite species in a group acts as an isomer and applying established techniques for isomer group thermodynamics [46]. The equation for standard transformed reactant Gibbs energy of formation, , in terms of metabolite species standard transformed Gibbs energy of formation, , is
| (1) |
where the summation is over all the metabolite species, j, within the pseudo-isomer group, i. Each metabolite species standard transformed Gibbs energy of formation is individually a function of ionic strength, pH and temperature. It is important to realize that Eq. 1 does not represent a linear average of the standard transformed Gibbs energy of formation of each metabolite species. In other words, reactant standard transformed Gibbs energy of formation is not additive in the standard transformed Gibbs energy of its metabolite species.
At specified ionic strength, pH and temperature, the mole fraction of a metabolite species, rj, with respect to its pseudo-isomer group, may be calculated using
| (2) |
where, again, the summation is over all the metabolite species, j, within a particular pseudo-isomer group, i [46]. This mole fraction rj, represents the fraction of a reactant which is present as a particular metabolite species j. It is evident from Eq. 2 that the mole fraction of a metabolite species is a non-linear function of its standard transformed Gibbs energy of formation. If each metabolite species in a pseudo-isomer group could be adjusted identically for temperature, ionic strength, and pH then it would be possible to adjust the properties of the reactant after it was approximated with the pseudo-isomer group approach. Mathematically, this follows since subtraction of any constant term from each in Eq. 2 has no effect on the mole fraction.
In vivo transformed reactant Gibbs energy
Transformed reactant Gibbs energy, , is given by , where is standard transformed reactant Gibbs energy and xi is the concentration of reactant i (Molar), which is the sum of the concentrations of its constituent metabolite species j. Quantitative metabolomic data [48], which can measure simultaneously a wide selection of reactant concentrations, is increasingly becoming available [49]. Quantitative metabolomic data can be used to estimate in vivo reactant transformed Gibbs energy [21]. Experimentally measured reactant concentrations indicate that the concentration of most amino acids [50], and reactants in central metabolism fall within the 0.02 – 20 mM range [21, 51]. Therefore, in the absence of metabolomic data for a particular reactant, we define the minimum and maximum standard transformed reactant Gibbs energy of formation, and , where we assume xmin = 0.02 mM and xmax = 20 mM. There are exception for water and dissolved gases (see Supplementary Material A.7).
Quantitative assignment of reaction directionality
Although, in principle, all microscopic processes in biochemistry are reversible, certain reactions are effectively irreversible at typical in vivo reactant concentrations. Let Sk ∈ ℧m,1 denote a column from a stoichiometric matrix [1], then the transformed Gibbs energy for a biochemical reaction is . In most modeling situations, metabolomic data will not be available for all reactants in a model. In this situation, we assume a physiological concentration range for each reactant. We define the minimum and maximum transformed Gibbs energy for a biochemical reaction with and , whereinf denotes the infimum, sup denotes the supremum, and reactant standard transformed Gibbs energy of formation is bounded . A reaction may be assigned to be quantitatively forward, if , or quantitatively reverse if . We assigned a reaction to be quantitatively reversible if the physiological range of biochemical reaction transformed Gibbs energy spans the zero line, i.e. and .
2.2 Physiological standard transformed reaction Gibbs energy
Given a set of reactions assigned to be quantitatively reversible, we now propose a metric to rank them in a descending sequence according to probability that each reaction is irreversible in the forward direction. Standard reaction Gibbs energy is defined for one molar concentration for each reactant, which far exceeds physiological concentration ranges in E. coli. We define a new physiological group contribution estimate for each reactant standard transformed Gibbs energy of formation,, where is the 1 Molar group contribution estimate of reactant standard Gibbs energy of formation, transformed to in vivo conditions, and xi,m is the geometric mean of the concentration range for each reactant, xi,m ≡ exp ((ln (xi,min) + ln (xi,max))/2). This refines the approach of Henry et al.[19], since xi,m represents a new standard concentration for each reactant, depending on its physiological range in vivo. We choose the arithmetic mean of minimum and maximum concentration since logarithmic concentration is linear in Gibbs energy.
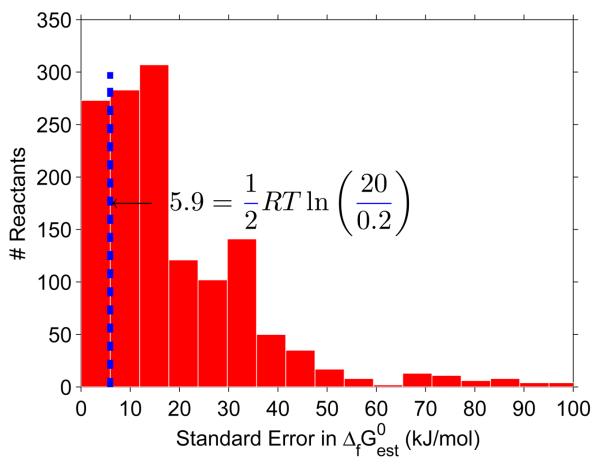
In the group contribution method, the standard error in estimated reactant standard Gibbs energy of formation, , assumes that the actual value of standard Gibbs energy of formation is normally distributed. Likewise, we assume that the actual value of physiological standard transformed reaction Gibbs energy, ΔrG′m, is also normally distributed. This is justified by comparing the relative magnitude of with RT ln (xi,max/xi,m) for reactants in iAF1260. Assuming a concentration range of 0.02 – 20 mM, the former is greater than the latter for 85% of reactants, see Figure 1. That is, for the majority of metabolites, the uncertainty in estimated reactant standard Gibbs energy of formation is more significant than the uncertainty associated with a lack of metabolomic data. The qualitatively reversible reactions, that are assigned to be quantitatively reversible, can be ranked based on the probability that the real physiological standard transformed reaction Gibbs energy is negative, P(ΔrG′m < 0). This probability that a reactions direction is forward is given by the cumulative of a normal distribution
| (3) |
where er f denotes the error function [52]. Ranking the reactions by the probability that physiological standard transformed reaction Gibbs energy is negative, P(ΔrG′m < 0), is superior to ranking reactions by mean estimated physiological standard transformed reaction Gibbs energy since the former accounts for the uncertainty in the group contribution method. (For the sake of clarity we omit a k subscript from P(ΔrG′m < 0), but it is specific to each reaction). Note that here probability is used in the Bayesian sense to reflect the confidence with which a directionality assignment can be made. If P(ΔrG′m < 0) = 0.5, then we have no confidence to assign direction either way.
Figure 1. Uncertainty in reactant Gibbs energy.
The majority of reactants in the E. coli iAF1260 model have a standard error in estimated standard Gibbs energy of formation which exceeds half the maximum error associated with assuming that reactant concentrations lie between 0.02 – 20 mM. This means that, for the majority of reactants, the uncertainty in reactant Gibbs energy is mostly from uncertainty in estimation of standard Gibbs energy, and not from uncertainty in concentration.
3 Results
3.1 Pseudo-isomer groups
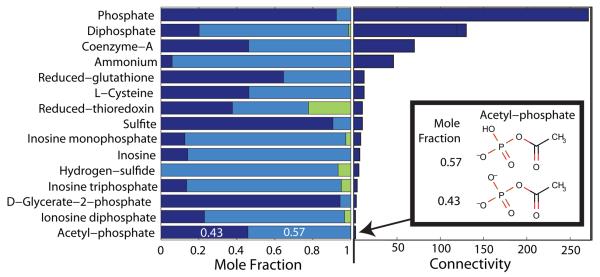
We applied quantitative assignment of reaction directionality to the genome scale model of E. coli metabolism, iAF1260 (1668 reactants, 2076 reactions) [28]. This highlighted a few pertinent methodological lessons which apply regardless of the organism of interest. The assumption that a metabolite is present as a single predominant metabolite species does not always apply. Figure 2 illustrates that certain reactants in E. coli do have significant mole fractions present as non-predominant metabolite species. For instance, acetyl-phosphate is almost equally present as C2H3O5P2− and C2H4O5P−, while a small mole fraction is predicted to be present as uncharged C2H5O5P. Each metabolite species has a different standard Gibbs energy of formation and responds individually to changes in pH, temperature and ionic strength. Where the necessary data was available, we calculated the standard transformed Gibbs energy of formation of each metabolite species as a function of pH, temperature and ionic strength. The standard transformed reactant Gibbs energy, and hence standard (Legendre) transformed reaction Gibbs energy is a function of the properties of each metabolite species in the respective pseudo-isomer groups, not just the predominant hydrogen ion dissociated form.
Figure 2. Non-predominant metabolite species.
At typical in vivo conditions, a minority of cytoplasmic reactants in E. coli (15/138 with available data), have significant mole fractions (> 0.05) present as non-predominant metabolite species. The length within each bar (left), apportioned to each color equals the mole fraction of the reactant present as a particular metabolite species. A bar with three colors indicates a reactant simultaneously present as three different metabolite species differing only in their state of protonation. Accurate prediction of standard transformed Gibbs energy of formation is especially important for reactants, such as Co-enzyme-A, which is almost equally present in the two different metabolites species forms, and participates as a reactant in many reactions (connectivity bar to the right). Another example is acetyl-phosphate which is also almost equally distributed, as C2H3O5P2− and C2H4O5P− (Inset). The mole fractions, corresponding to a pH 7.7 [43, 44], ionic strength 0.25 M and temperature 310.15 K, were calculated as described in Section 2.1.
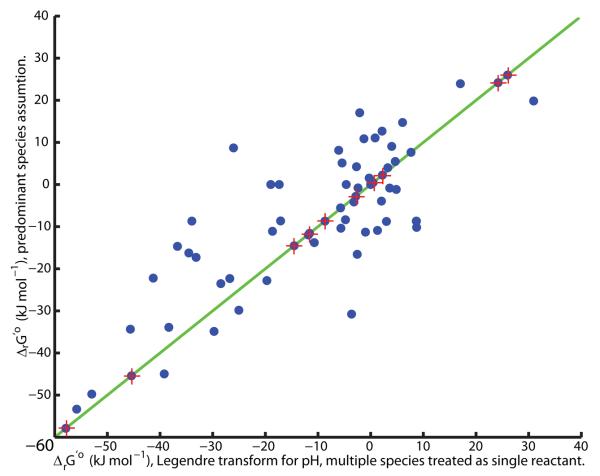
Another approach reported in the literature assumes that all reactants can be approximated by a single predominant metabolite species at a given pH [19, 20]. There, in order to represent a known constant hydrogen ion concentration, [H+], a new standard hydrogen ion Gibbs energy of formation is defined, ΔfGo(H+) ≡ RT ln [H+], and the Legendre transformation of all metabolite species at constant pH is omitted [19, 20]. Thereafter, standard ’transformed’ reaction Gibbs energy is calculated with the newly defined standard hydrogen ion Gibbs energy of formation and standard Gibbs energy of formation for the remaining reactants, each approximated by a single metabolite species. Figure 3 illustrates that there is a significant difference between the standard (Legendre) transformed reaction Gibbs energy and the standard ’transformed’ Gibbs energy of the same reaction, calculated when one assumes that each reactant can be approximated with one predominant metabolite species. As expected, in exceptional cases, where all reactants involved in a biochemical reaction exist as one metabolite species, the standard ’transformed’ Gibbs energy of a reaction is identical to the standard transformed Gibbs energy of the same reaction.
Figure 3. The importance of a true Legendre Transform.
When cytoplasmic pH is held constant by buffering, a Legendre transform of metabolite species standard Gibbs energy of formation defines a standard transformed Gibbs energy of formation for each metabolite species. Reactant standard transformed Gibbs energy of formation, and therefore reaction standard transformed Gibbs energy, depends non-additively on the standard transformed metabolite species Gibbs energy of formation. Due to this non-additivity, when reactants are present as multiple species, differing in their state of protonation, it is erroneous to replace transformation of metabolite species standard Gibbs energy of formation, with adjustment to the hydrogen ion standard Gibbs energy of formation, when calculating reaction standard transformed Gibbs energy. The latter gives rise to an erroneous estimate of standard ‘transformed’ reaction Gibbs energy, as illustrated above for the reactions in the central metabolic E. coli model [61] at pH 7.7, zero ionic strength, atmospheric pressure and temperature 298.15K. The exception is when each reactant involved in a reaction is present as only one metabolite species. In this case both approaches agree (stars on the diagonal).
3.2 Qualitative versus quantitative reaction directionality in E. coli
To quantitatively assign reaction directionality, where available, we used standard reactant Gibbs energy of formation back-calculated from experimentally measured equilibrium constants in preference to group contribution estimates. However, a minority of the reactants in E. coli (311/1668) have standard Gibbs energy of formation back-calculated from experimentally measured equilibrium constants [30]. Most of the remaining reactant standard Gibbs energy of formation (1127/1668) can be estimated using group contribution methodology [31]. Certain reactants have no estimated standard Gibbs energy of formation due to structural subgroups in metabolite species not covered by the current version of this method (230/1668 or 163/1039 unique iAF1260 reactants).
We compared quantitative assignment of reaction directionality with qualitatively assigned reaction reconstruction directionality in the genome scale metabolic model of E. coli, iAF1260 [28]. The heuristics for the latter are given in the Supplementary Material A.8. Overall, quantitative assignment of reaction directionality results in a relaxation of qualitative directionality constraints, see Table 1. Directionality changes are in three categories: i) unchanged directionality ii) relative relaxation of reaction directionality (qualitatively forward yet quantitatively reversible), and iii) tightened or reversed reaction directionality. Unless otherwise specified, we refer to quantitative assignment at a temperature of 310.15 K, ionic strength of 0.25 M, pH of 7.7 [43, 44], and an accommodating physiological concentration range of 0.02 – 20 mM. Initially, we assumed thermodynamic reversibility for internal reactions (309/2077) where data is missing for at least one reactant.
Table 1.
Comparison of qualitative versus quantitative assignment of reaction directionality for the genome scale constraint-based model of E. coli, iAF1260 [28]. Quantitative assignments were calculated at specific temperature, ionic strength and pH. Each cell represents the number of reactions with a particular type of change in directionality assignment. Reaction directionality was calculated with metabolite group contribution estimates [31] alone (right) or with both group contribution estimates and reactant standard transformed Gibbs energy of formation back calculated from experimentally measured equilibrium constants, when available (left) [30]. By incorporating the uncertainty associated with group contribution estimates (), reaction directionality is considerably relaxed.
| Qualitative directionality assignment |
Quantitative directionality assignment |
only | |||||
|---|---|---|---|---|---|---|---|
| Forward | Reversible | 603♠ (103, 376, 124) |
1011 (103, 774, 134) |
612 (89, 399, 89) |
609 (0, 406, 203) |
1133 (0, 853, 280) |
642 (0, 440, 202) |
| Reversible | Forward | 10 | 6 | 8 | 7 | 1 | 7 |
| Reversible | Reverse | 3 | 1 | 2 | 11 | 0 | 11 |
| Forward | Reverse | 11 | 5♣ | 12 | 57 | 35 | 56 |
| Temp. (K) | 310.15 | 310.15 | 298.15 | 310.15 | 310.15 | 298.15 | |
| Ionic strength (M) |
0.25 | 0.25 | 0 | 0.25 | 0.25 | 0 | |
| pH | 7.7 | 7.7 | 7 | 7.7 | 7.7 | 7 | |
| 0 | 1 | 0 | 0 | 1 | 0 | ||
The lowest number of reactions with reversal of directionality, i.e. forward irreversible changed to reverse irreversible, is achieved when using the following conditions: (i) use of reactant standard transformed Gibbs energy of formation back calculated from experimentally measured equilibrium constants, (ii) incorporation of uncertainty associated with group contribution estimates.
The numbers of qualitatively forward, quantitatively reverse reactions are further decomposed depending on the provenance of the metabolite data (, , ).
3.3 Quantitative relaxation of qualitatively assigned reaction directionality
In iAF1260, most of the internal reactions (1524/2077) have been qualitatively assigned to be irreversible in the forward direction. Of these forward reactions, about a third are also quantitatively forward (638/1524). Another third of qualitatively forward reactions are assigned to be quantitatively reversible (603/1524), see Table 1. Taking into account the uncertainty in group contribution estimation of metabolite species standard Gibbs energy of formation, the fraction of qualitatively forward reactions changed to quantitatively reversible almost doubles (1011/1524). Relaxation of directionality constraints for so many reactions would significantly increase the flexibility of the resulting iAF1260 constraint-based model. Such relaxation significantly effects model predictions, therefore, we conducted a detailed analysis of these qualitatively forward, yet quantitatively reverse reactions.
Quantitative relaxation of reaction directions with no uncertainty in ΔrG′0
A tenth of qualitatively forward yet quantitatively reversible reactions (103/1011) exclusively involved reactant standard transformed Gibbs energy of formation back calculated from equilibrium constants. The majority of these reactions do indeed have a negative standard transformed reaction Gibbs energy, but within our accommodating physiological range of reactant concentration, they are quantitatively reversible (See Supplementary Figure 10). Strictly, these reactions are quantitatively reversible, i.e., and , but most are probably forward at in vivo concentrations.
Many of the quantitatively reversible reactions transport metabolites between compartments. Although the net electrical charge within a compartment is assumed to be neutral, an electrical potential difference can exist between compartments. In E. coli, the cytoplasmic side of the cytoplasmic membrane is more negative than the periplasmic side [53]. A thorough treatment of electrochemical potential difference is beyond the scope of this current work. However, even when cytoplasm and periplasm are assumed to have an identical pH, an electrical potential difference still exists across the cytoplasmic membrane. We assumed an electrical potential difference of 90 mV, which corresponds to a Gibbs energy change of −8.7 kJ mol−1 per negative charge transported from cytoplasm to periplasm [53]. Therefore, the passive transport of negatively charged reactants, e.g., formate, from cytoplasm to periplasm is thermodynamically favorable at equal concentrations of formate in both compartments. However, if the concentration of formate is high in the periplasm, and low in the cytoplasm, this can reverse the direction of formate diffusion. Therefore, many transport reactions are quantitatively assigned to be reversible within a physiological range of reactant concentration. This is the case regardless of the provenience of the reactant standard Gibbs energy of formation, see Supplementary Figure 11.
Quantitative relaxation of reaction directions with uncertainty in metabolite
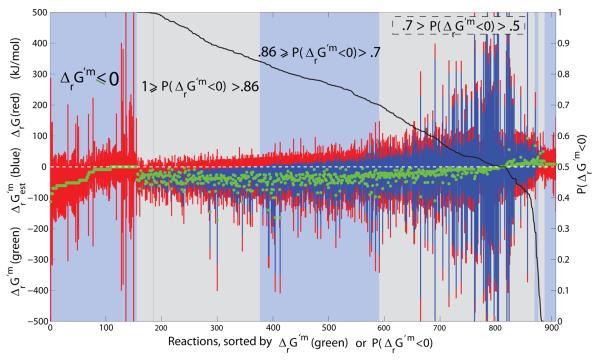
A small minority (134/1011) of the qualitatively forward yet quantitatively reversible reactions are assigned based solely on group contribution estimates of reactant standard transformed Gibbs energy of formation. The uncertainty in these estimates combines with variability in metabolite concentration to increase the feasible range of reaction Gibbs energy. The remaining majority (774/1011) of the qualitatively forward yet quantitatively reversible assignments are based on standard transformed Gibbs energy of formation for reactants estimated by the group contribution method combined with standard transformed Gibbs energy of formation for reactants back calculated from equilibrium constants. The latter reactants include most of the widely used co-enzymes [54] and prosthetic groups. In order to understand the reasons for relaxation of so many reaction directions ((134+774)/1011), we categorized the reactions. Figure 4 illustrates all reactions that are qualitatively forward yet quantitatively reversible (908/1011), using at least one group contribution estimate of reactant standard transformed Gibbs energy of formation. Apart from transport reactions, the reactions in Figure 4 are ordered by descending probability of the reaction being forward, P(ΔrG′m < 0), at physiological standard concentrations of reactants.
Figure 4. Qualitatively forward, quantitatively reverse reactions.
The reactions that are qualitatively assigned to be forward in iAF1260, yet quantitatively reversible, using at least one group contribution estimate of reactant standard transformed Gibbs energy of formation. The feasible range of ΔrG′ and are given as red and blue bars respectively. Far left are transport reactions with negative or zero physiological standard transformed reaction Gibbs energy, ΔrG′m ≤ 0, but reversible depending on concentration of reactants. Reactions with uncertainty due to estimation of standard transformed reaction Gibbs energy are rank ordered by decreasing probability that physiological standard transformed reaction Gibbs energy is negative. In mathematical notation this probability is represented by the symbol P(ΔrG′m < 0), as defined in Eq. 3, and used in situabove to denote the intervals as follows: Reactions with P(ΔrG′m < 0) > 0.7 were assumed to be irreversible in the forward direction, and reactions with P(ΔrG′m < 0) < 0.3 or ΔrG′m > 0, were assumed to be irreversible in the reverse direction. See Figure 12 for a detailed illustration of the latter reactions. Reactions with 0.7 ≥ P(ΔrG′m < 0) > 0.3 were allowed to be quantitatively reversible in lieu of the large uncertainty in estimation of standard transformed reaction Gibbs energy.
Thermodynamically favorable, qualitatively forward transport reactions
For most transport reactions, there is no uncertainty in physiological standard transformed reaction Gibbs energy since the same reactant is present on both sides of the reaction. In Figure 4, transport reactions are sorted by ascending physiological standard transformed reaction Gibbs energy since P(ΔrG′m < 0) is undefined if the denominator in Eq. 3 is zero. A non-zero standard transformed reaction Gibbs energy for a transport reaction is due to net transport of a charged metabolite between the cytoplasm and periplasm. Of the qualitatively forward yet quantitatively reversible reactions, 155/1011 result in net transport of a negative (or neutral) charge from the cytoplasm to the periplasm and therefore have a negative (or zero) physiological standard transformed reaction Gibbs energy, ΔrG′m ≤ 0. These transport reactions are qualitatively assigned to be forward, but within the physiological range of reactant concentration, they are quantitatively reversible.
Thermodynamically unfavorable, qualitatively forward transport reactions
The forward direction of a transport reaction, as for any reaction, is defined by convention. Assuming the concentration range of the reactant is the same in both compartments, reactions which transport a positively charged reactant from the cytoplasm to the periplasm have a positive physiological standard transformed reaction Gibbs energy, ΔrG′m > 0. Likewise, reactions which transport a negatively charged reactant from periplasm to the cytoplasm have a positive standard transformed reaction Gibbs energy. At equal cytoplasmic and periplasmic reactant concentrations these reactions would operate in reverse, despite being qualitatively assigned to be forward reactions. These 21 transport reactions are detailed in Supplementary Figure 12. Any facilitated diffusion reaction in this group would have to have a higher periplasmic than cytoplasmic concentration in order to proceed in the forward direction as qualitatively assigned, e.g., undecaprenyl phosphate transport cytoplasm to periplasm.
At an equal pH of cytoplasm and periplasm, any proton symport reaction, transporting a reactant with a charge of −2 into the cell, would require more than one proton to translocate across the periplasmic membrane in order to make the reaction thermodynamically favorable. It may be that the proton stoichiometry of these reactions needs to be revised in order to take this into account. For example the reaction ‘GlcNAc anhMurNAc tetrapeptide transport in via proton symport’, which symports anhMurNAc tetrapeptide (N-Acetyl-D-glucosamine (anhydrous) N-Acetylmuramyl-tetrapeptide), a biopolymer in the bacterial cell wall, with a charge of −2, from the periplasm to the cytoplasm, see Supplementary Figure 12. This result indicates how thermodynamic consideration of reaction directionality can highlight reactions with thermodynamically infeasible stoichiometry.
Qualitatively forward reactions, quantitatively reversible within a physiological concentration range
The probability that physiological standard transformed reaction Gibbs energy less than zero, P(ΔrG′m < 0), Eq. 3, attempts to quantify the probability that a reaction is indeed irreversible in the forward direction at physiological standard concentrations of reactants. Reactions that are most probable to be irreversible in the forward direction are those where the physiological standard transformed reaction Gibbs energy is negative, even one standard deviation from the mean, i.e. 1 ≥ P(ΔrG′m < 0) ≥ 0.86. Of the qualitatively forward, yet quantitatively reversible reactions, a quarter (221/1011) are probably forward by this reasoning. By the same reasoning for the opposite direction, 0.14 ≥ P(ΔrG′m < 0) ≥ 0, there are 11 reactions that are probably reverse, but are qualitatively assigned to operate in the forward direction, see Supplementary Figure 12. These quantitative reaction directionality assignments are interesting because they possibly change the direction of qualitative assignment.
The intermediate range of probability of forward physiological standard transformed reaction Gibbs energy, 0.86 ≥ P(ΔrG′m < 0) > 0.14, still accounts for half (500/1011) of the qualitatively forward, yet quantitatively reverse reactions. Figure 4 illustrates that a large uncertainty in physiological standard transformed reaction Gibbs energy precludes definitive quantitative forward assignment for many of these reactions. We arbitrarily set the 214 reactions with 0.86 ≥ P(ΔrG′m < 0) > 0.7 to be quantitatively forward, the 279 reactions with 0.7 ≥ P(ΔrG′m < 0) > 0.3 to be quantitatively reversible and the remaining 7 reactions with 0.3 ≥ P(ΔrG′m < 0) > 0.14 to be quantitatively reverse.
Whatever upper cutoff is chosen for qualitatively forward reactions, then in order to be logically consistent, one minus the same cutoff must be used to quantitatively assign other reactions to be reverse. Our conservative upper & lower cutoff was chosen to compensate for the uncertainty in group contribution estimates of reactant standard Gibbs energy. Even with these conservative cutoffs, the resulting model cannot produce biomass in glucose minimal medium, because certain qualitatively forward reactions are essential in this direction and therefore cannot be reversed. Computational analysis can reliably predict, which reaction directions are essential in vivo for a given boundary condition [8, 55]. We identified the 7 reactions that were essential for production of biomass in glucose minimal medium, yet seemed thermodynamically unfavorable according to 0.3 ≥ P(ΔrG′m < 0) (See Supplementary Figure 12). This highlights the importance of incorporating metabolite concentration data to refine thermodynamic assignment of reaction directionality [21] since it is ultimately the transformed Gibbs energy of a biochemical reaction, ΔrG′, that determines directionality.
L-glutamate and reaction directionality
The amino acid L-glutamate is the major nitrogen donor in the cell, distributing ~88% of the total nitrogen that ends up in biomass, largely via transamination reactions. It has been measured at a relatively high~ 100 mM concentration in a range of growth conditions [56]. L-glutamate is a reactant in three of the qualitatively forward reactions, that could possibly be assigned reverse based on consideration of physiological standard transformed reaction Gibbs energy alone (Supplementary Figure 12). L-glutamate is a substrate for glutamate 5 kinase (E.C. 2.7.2.11) and leucine transaminase (E.C. 2.6.1.6), yet a product of the reaction catalyzed by 1-pyrroline 5-carboxylate dehydrogenase (EC 1.5.1.12). The reaction catalyzed by leucine transaminase is essential for production of biomass from glucose minimal medium in silico and in vivo [57]. A high concentration of L-glutamate favors the forward direction of this reaction. In the same conditions, the other two reactions involving L-glutamate are not essential in silico.
Leucine transaminase catalyzes the transfer of the amine group in L-glutamate to 4-methyl-2-oxopentanoate to form L-leucine. This reaction can also be catalyzed by branched-chain-amino-acid transaminase (EC 2.6.1.42) which catalyses the first step in the catabolism of branched-chain amino acids, being leucine, isoleucine and valine. At 298.15 K, pH 7.21, and ionic strength 0.31 mol kg−1, the apparent equilibrium constant of
was reported to be 2.42 ± 0.25 with respect to the forward direction [58]. Without error this corresponds to a standard transformed Gibbs energy of −2.19 kJ mol−1. This indicates that the reverse direction, biosynthesis of L-leucine, is thermodynamically unfavorable at equal concentrations of substrates and products. This indicates that the high concentration of glutamate observed in the cell may well be thermodynamically necessary to drive the biosynthesis of L-leucine. On the contrary, depending on the needs of the cell, all else being equal, the reaction will run in the opposite direction at low glutamate concentration. This illustrates how thermodynamic data can be used to interpret the meaning of metabolomic data [21].
3.4 Quantitative tightening of qualitatively assigned reaction directionality
Using quantitative assignment, a small minority of reaction directions are tightened or reversed, depending on the quantitative method used. Using group contribution estimates alone, there are at least 35 qualitatively reversible reactions that are deemed to be quantitatively irreversible, see Table 1. This number rises to 56 reactions when the uncertainty associated with the group contribution methodology is not taken into account. In contrast, the use of standard Gibbs energy of formation back calculated from experimentally measured equilibrium constants, in place of group contribution estimates, reduces the number of reaction directions that are tightened or reversed. In particular, of 553 qualitatively reversible internal reactions in iAF1260, only 6 are deemed to be quantitatively irreversible using standard transformed Gibbs energy of formation back calculated from experimentally measured equilibrium constants, in preference to group contribution estimates. Of course, group contribution estimates are still required for the majority of reactants. Of 1524 qualitatively forward reactions, only 5 are deemed to be quantitatively irreversible in the reverse direction.
The tightened or reversed directionality assignments, directly conflict with qualitative assignments and therefore we investigated the literature on biochemical and/or thermodynamic characterisation experiments for each of these reactions. The detailed results of this study are given in Supplementary Material A.9. Quantitative tightening of reaction directionality was supported for a minority of reactions, e.g. D-Erythrose-4-phosphate dehydrogenase reaction. Other reactions had experimental evidence indicating that quantitative assignment was too tight, which may perhaps be improved by more accurate modeling of the effect of of Magnesium on the standard transformed Gibbs energy of ATP, e.g. galactokinase reaction. As discussed by Alberty [29], the concept of a pseudo-isomer group may be extended to different metal ion dissociated forms, e.g., when pMg = −log10[Mg2+] is a known constant. The group contribution method seemed to have difficulty in estimating the standard Gibbs energy of metabolites with an imidazole ring structure, giving rise to erroneous, over tight assignments for certain reactions, e.g. IMP cyclohydrolase. Similarly, certain qualitatively forward reactions were also incorrectly predicted to be quantitatively reverse for reactions involving other metabolites with complex structure, e.g. glycogen. Ongoing refinements to the group contribution method can be expected to iron out these issues with structurally complex metabolite species[31].
3.5 Quantitative assignment of reaction directionality and biomass production in E. coli
Using the stoichiometry of the latest genome scale metabolic model, iAF1260[28], we quantitatively set reaction directionality according to local thermodynamic constraints and compared growth rate prediction against those experimentally measured. A first pass thermodynamic assignment of reaction directionality based on ΔrG′ alone, resulted in a minority of reactions being assigned thermodynamically forward (242/2077), and a few reactions being thermodynamically reverse (6/2077). A few of these reversed reactions were incorrectly quantitatively assigned, essential for growth, and therefore qualitatively set to forward, ignoring the thermodynamic assignment. Using flux balance analysis [33, 59] to maximize reaction representing the production of biomass, at first, the biomass production rate of the resulting model far exceeded the in vivo growth rate on glucose minimal medium. This indicates that significant reaction directionality constraints were missing since the published iAF1260 model does reproduce experimentally reported growth rates in various media.
At this point, we attempted to identify the minimum number of biochemically reasonable qualitative assignments necessary in order to match the growth rate observed in vivo. The qualitative assignments were based the on the experimental biochemical literature used to assign reaction directionality in the iAF1260 reconstruction [28]. In addition, care was taken to qualitatively assess prediction of internal fluxes, such as flux through the electron transport chain, as expected during aerobic growth. First, we assigned the qualitative reconstruction directions to reactions without any thermodynamic data (310/2011). Similarly for reactions involving a quinone, since quinones are structurally complex metabolite species with significant uncertainty in group contribution estimates. Then, using the probabilistic criteria discussed above, we made these further quantitative assignments: P(ΔrG′m < 0) > 0.7 ⇔forward, 0.7 ≥ P(ΔrG′m < 0) > 0.3 ⇔reversible, and 0.3 ≥ P(ΔrG′m < 0) ⇔reverse, except for in silico essential reactions.
In addition, it was necessary to eliminate artefactual excess ATP synthesis due to (i) reversal of ATP consuming ATP-binding cassette transport reactions, and (ii) net shuttling of protons from the cytoplasm into the periplasmic compartment via transport reactions, leading to excess ATP synthase flux. We assigned reconstruction directions to all transport reactions involving proton symport or antiport, and all ATP-binding cassette transport reactions. The majority of transport reactions qualitatively assigned a forward direction in this way do indeed have a negative physiological standard transformed reaction Gibbs energy, see Supplementary Figure 11. For transport reactions, compartmentally resolved reactant concentration, if available, would significantly influence reaction directionality, since, for many such reactions, the standard Gibbs energy is close to zero.
We qualitatively assigned reaction directionality to cytoplasmic reactions involving the cofactors ATP, GTP, CTP & UTP. In addition, we incorporated experimentally reported concentration ranges for key cofactors (ATP, ADP, AMP, NAD, NADH, NADP, NADPH) measured for E. coli growing aerobically on glucose minimal medium [51]. At glucose and oxygen uptake rates of 11 and 18.2 mmol g−1 h−1 respectively, the experimentally observed growth rate is 0.82 h−1 [60]. The aforementioned qualitative reaction directionality assignments, which override the quantitative assignments, reduce the growth rate to 1.1 h−1, which compares favorably with that observed experimentally. At this margin of error, setting the effective stoichiometry of oxidative phosphorylation by constraints on the relative fluxes through various NADH dehydrogenases and terminal oxidases has a significant effect on growth rate [28]. It is clear that the latter modeling issue awaits a thorough investigation. In summary, quantitative assignment of reaction directionality must be accompanied by qualitative assignment, for certain classes of reactions, in order to match experimental data with constraint based models of metabolism.
4 Discussion
In principle, the application of the second law of thermodynamics to metabolic reactions results in a constraint on the direction of all network reactions. However, this assumes that all reactant concentrations and standard Gibbs energies are are known. Using experimentally measured apparent equilibrium constants, Alberty has published tables of standard transformed reactant Gibbs energies for 200 reactants [29, 30]. In addition, group contribution estimates of standard reactant Gibbs energy have recently become available for two thirds of the 15,000 reactants in the KEGG compound database [31]. We have integrated these estimates, with Alberty's tables in order to quantitatively constrain the directions in the E. coli genome scale metabolic model.
Standard Gibbs energy applies to chemical standard conditions, which are markedly different from in vivo conditions. Therefore, following the methods developed by Alberty[29], we have theoretically summarized and practically implemented the main steps in transformation of chemical standard thermodynamic potentials into biochemical standard thermodynamic potentials. The large buffering capacity of proteins, relative to the size of the soluble reactant pool means that pH is effectively held constant. Therefore, we use the transformed reaction Gibbs energy to provide the criterion for spontaneous change, and therefore the direction of net flux, at in vivo temperature, pressure, pH and ionic strength.
Even in the absence of quantitative metabolomic data, thermodynamic assignment of reaction directionality can be made by assuming an accommodating range of reactant concentration (0.02 – 20 mM). In E. coli, our thermodynamic assignment of reaction directionality rarely conflicts with qualitatively assigned reconstruction direction based on experimental literature. This indicates that the same algorithms may be reliably used to constrain reaction directionality for other organisms without the same breadth of experimental evidence as exists for E. coli. However, a large proportion of reaction directions, which were qualitatively assigned to be irreversible in E. coli, become quantitatively reversible, by transformed reaction Gibbs energy alone. Partially, this is due to the accommodating range of reactant concentration, especially for transport reactions. However, the main reason is that the uncertainty associated with computational estimation of standard reactant Gibbs energy can give rise to a significant uncertainty in standard transformed reaction Gibbs energy. This is especially so when structurally complex reactants are involved.
We partially overcame the uncertainty issue by formulating a probabilistic metric expressing our confidence that a given reaction is irreversible, given the uncertainty in standard reactant Gibbs energy. Using this Bayesian probabilistic metric, it is possible to quantitatively assign a large number of reactions to be irreversible, at a given confidence cutoff. Even still, some qualitative assignment of reaction directionality is necessary in order to sufficiently constrain the genome scale E. coli model to agree with experimentally observed growth rate in glucose minimal medium. In particular, this is so for quinone coupled oxidative phosphorylation reactions transport reactions, and reactions involving nucleotide cofactors.
Quantitative metabolomics data, combined with standard transformed reactant Gibbs energy calculations, would increase the number of predicted effectively irreversible reactions. However, metabolite concentration data only becomes important for determining reaction directionality when the uncertainty in group contribution estimation of reactant Gibbs energy is low (⪅ 6 kJ mol−1). Cofactor pair concentration ratios are particularly useful since these reactants are involved in such a large proportion of reactions and since experimentally derived standard reactant Gibbs energy are available for many cofactors from the tables compiled by Alberty [29]. Beyond cofactors, the concentration of other key metabolites are also important for determining the direction of essential anabolic reactions. For instance, we provide evidence that the high concentration of cytoplasmic L-glutamate, observed in E. coli [56], may well be thermodynamically necessary to drive the biosynthesis of L-leucine.
We observed that reaction directionality is relatively insensitive to the specific adjustments for temperature, pH and ionic strength necessary to represent in vivo conditions. This is more due to the uncertainty in standard reactant Gibbs energy than the absence of metabolomic data. Integration of standard reactant Gibbs energy back calculated from equilibrium constants significantly reduces the number of false positive irreversible reactions, as compared to the use of group contribution estimates alone. Nevertheless, group contribution estimates are essential to match the broad scope of current genome scale models, therefore it is evident that these approaches to the thermodynamics of biochemical reactions are complementary.
Supplementary Material
Acknowledgements
This work was supported by NIH grant Grant 5R01GM057089-11 and a National University of Ireland, Galway, Science Faculty Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palsson BØ. Systems Biology: Properties of Reconstructed Networks. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 2.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2009 doi: 10.1038/nprot.2009.203. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pál C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440(7084):667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 4.Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5(4):264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14(2):301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett CL, Herring CD, Reed JL, Palsson BØ. The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states. Proc Natl Acad Sci U S A. 2005;102(52):19103–19108. doi: 10.1073/pnas.0505231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samal A, Jain S. The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response. BMC Syst Biol. 2008;2:21. doi: 10.1186/1752-0509-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards JS, Ibarra RU, Palsson BØ. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19(2):125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 9.Ibarra RU, Edwards JS, Palsson BØ. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420(6912):186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 10.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BØ. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429(6987):92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 11.Herrgård MJ, Fong SS, Palsson BØ. Identification of genome-scale metabolic network models using experimentally measured flux profiles. PLoS Comp Bio. 2006;2(7):e72. doi: 10.1371/journal.pcbi.0020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BØ. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103(46):17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 14.Patil KR, Rocha I, Förster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BioMed Central Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci U S A. 2007;104(19):7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feist AM, Palsson BØ. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26(6):659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savinell JM, Palsson BO. Network analysis of intermediary metabolism using linear optimization. i. development of mathematical formalism. J Theor Biol. 1992;154(4):421–454. doi: 10.1016/s0022-5193(05)80161-4. [DOI] [PubMed] [Google Scholar]
- 18.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:e119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90(4):1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92(5):1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kümmel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol Syst Biol. 2006;2:e34. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kümmel A, Panke S, Heinemann M. Systematic assignment of thermodynamic constraints in metabolic network models. BioMed Central Bioinformatics. 2006;7(1):512. doi: 10.1186/1471-2105-7-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberty RA. A short history of the thermodynamics of enzyme-catalyzed reactions. J Biol Chem. 2004;279(27):27831–27836. doi: 10.1074/jbc.X400003200. [DOI] [PubMed] [Google Scholar]
- 24.Burton K, Krebs HA, Kornberg HL. Energy Transformations in Living Matter. Springer-Verlag; Berlin: 1957. [PubMed] [Google Scholar]
- 25.Yang F, Qian H, Beard DA. Ab initio prediction of thermodynamically feasible reaction directions from biochemical network stoichiometry. Metab Eng. 2005;7(4):251–259. doi: 10.1016/j.ymben.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Yang Feng, Beard Daniel A. Thermodynamically based profiling of drug metabolism and drug-drug metabolic interactions: a case study of acetaminophen and ethanol toxic interaction. Biophys Chem. 2006;120(2):121–134. doi: 10.1016/j.bpc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Heuett WJ, Qian H. Combining flux and energy balance analysis to model large-scale biochemical networks. J Bioinform Comput Biol. 2006;4(6):1227–1243. doi: 10.1142/s0219720006002430. [DOI] [PubMed] [Google Scholar]
- 28.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3(1):e121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alberty RA. Thermodynamics of Biochemical Reactions. Wiley-Interscience; Hoboken, NJ: 2003. [Google Scholar]
- 30.Alberty RA. Biochemical Thermodynamics: Applications of Mathematica. Wiley-Interscience; Hoboken, NJ: 2006. [PubMed] [Google Scholar]
- 31.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys J. 2008;95(3):1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavrovouniotis ML. Estimation of standard Gibbs energy changes of biotransformations. J Biol Chem. 1991;266(22):14440–14445. [PubMed] [Google Scholar]
- 33.Varma A, Palsson BØ. Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. J Theor Biol. 1993;165(4):477–502. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 34.Varma A, Palsson BØ. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol. 1994;60(10):3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross J. Thermodynamics and fluctuations far from equilibrium, volume 80 of Springer Series in Chemical Physics. Springer; New York: 2008. [Google Scholar]
- 36.Schnell S, Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Prog Biophys Mol Biol. 2004;85(23):235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Minton AP. How can biochemical reactions within cells differ from those in test tubes? J Cell Sci. 2006;119(14):2863. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 38.Verkman AS. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem Sci. 2002;27(1):27–33. doi: 10.1016/s0968-0004(01)02003-5. [DOI] [PubMed] [Google Scholar]
- 39.Kao HP, Abney JR, Verkman AS. Determinants of the translational mobility of a small solute in cell cytoplasm. J Cell Biol. 1993;120(1):175–184. doi: 10.1083/jcb.120.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biemans-Oldehinkel Esther, Mahmood Nik A B N, Poolman Bert. A sensor for intracellular ionic strength. Proc Natl Acad Sci U S A. 2006;103(28):10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dill KA, Bromberg S. Molecular driving forces: Statistical thermodynamics in Chemistry and Biology. Garland Science; London: 2003. [Google Scholar]
- 42.Neidhardt FC, Curtis R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology Press; Washington, DC: 1996. [Google Scholar]
- 43.Padan E, Schuldiner S. Intracellular pH and membrane potential as regulators in the prokaryotic cell. J Membr Biol. 1987;95(3):189–198. doi: 10.1007/BF01869481. [DOI] [PubMed] [Google Scholar]
- 44.Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189(15):5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slonczewski JL, Macnab RM, Alger JR, Castle AM. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J Bacteriol. 1982;152(1):384–399. doi: 10.1128/jb.152.1.384-399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith WR, Missen RW. Chemical Reaction Equilibrium Analysis: Theory and Algorithms. Wiley; New York: 1982. [Google Scholar]
- 47.Fersht A. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding. W. H. Freeman; New York: 1999. [Google Scholar]
- 48.Fiehn O. Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(12):155–171. [PubMed] [Google Scholar]
- 49.Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci U S A. 2006;103(51):19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita M. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316(5824):593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 51.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5(8):593–9. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gowers T, Barrow-Green J, Leade I. The Princeton Companion to Mathematics. Princeton University Pres; Princeton: 2008. [Google Scholar]
- 53.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186(18):6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alberty RA. Standard transformed Gibbs energies of coenzyme A derivatives as functions of pH and ionic strength. Biophys Chem. 2003;104(1):327–334. doi: 10.1016/s0301-4622(02)00390-3. [DOI] [PubMed] [Google Scholar]
- 55.Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BØ, Agarwalla S. Experimental and computational assessment of conditionally essential genes in Escherichia coli? J Bacteriol. 2006;188(23):8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol. 2006;2(10):529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- 57.Umbarger HE. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:532–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 58.Tewari YB, Goldberg RN, Rozzell JD. Thermodynamics of reactions catalysed by branched-chain-amino-acid transaminase. J Chem Thermodyn. 2000;32(10):1381–1398. [Google Scholar]
- 59.Varma A, Palsson BØ. Metabolic capabilities of Escherichia coli: II. Optimal growth patterns. J Theor Biol. 1993;165(4):503–522. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 60.Fischer E, Zamboni N, Sauer U. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13c constraints. Anal Biochem. 2004;325(2):308–316. doi: 10.1016/j.ab.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 61.Orth JD, Fleming RMT, Ø Bernhard. Escherichia coli and Salmonella: Cellular and Molecular Biology, chapter Reconstruction and use of microbial metabolic networks: the core Escherichia coli metabolic model as an educational guide (accepted) ASM Press; 2009. Palsson. [DOI] [PubMed] [Google Scholar]
- 62.Gibbs JW. Elementary principles in statistical mechanics, developed with especial reference to the rational foundation of thermodynamics. Dover Publications; New York: 1902. [Google Scholar]
- 63.Baierlein R. The elusive chemical potential. Am J Phys. 2001;69(4):423–434. [Google Scholar]
- 64.Planck M. Treatise on Thermodynamics. Courier Dover Publications; Chelmsford, MA: 1945. [Google Scholar]
- 65.Alberty RA. IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN). Recommendations for nomenclature and tables in biochemical thermodynamics. Recommendations 1994. Eur J Biochem. 1996;240(1):1–14. doi: 10.1111/j.1432-1033.1996.0001h.x. [DOI] [PubMed] [Google Scholar]
- 66.Berry SR, Rice SA, Ross J. Physical Chemistry. 2nd edition Oxford University Press; Oxford: 2000. [Google Scholar]
- 67.Alberty RA. Legendre transforms in chemical thermodynamics. J Chem Thermodyn. 1997;29(5):501–516. [Google Scholar]
- 68.Alberty Robert A. The role of water in the thermodynamics of dilute aqueous solutions. Biophys Chem. 2003;100(13):183–192. doi: 10.1016/s0301-4622(02)00280-6. [DOI] [PubMed] [Google Scholar]
- 69.Zhao G, Pease AJ, Bharani N, Winkler ME. Biochemical characterization of gapb-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli k-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J Bacteriol. 1995;177(10):2804–2812. doi: 10.1128/jb.177.10.2804-2812.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boschi-Muller S, Azza S, Pollastro D, Corbier C, Branlant G. Comparative enzymatic properties of gapb-encoded erythrose-4-phosphate dehydrogenase of escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1997;272(24):15106–15112. doi: 10.1074/jbc.272.24.15106. [DOI] [PubMed] [Google Scholar]
- 71.Atkinson MR, Johnson E, Morton RK. Equilibrium constant of the galactokinase reaction and free energy of hydrolysis of adenosine triphosphate. Nature. 1959;184:1925–1927. doi: 10.1038/1841925a0. [DOI] [PubMed] [Google Scholar]
- 72.Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, Suzuki H. A novel putrescine utilization pathway involves gamma-glutamylated intermediates of Escherichia coli k-12. J Biol Chem. 2005;280(6):4602–4608. doi: 10.1074/jbc.M411114200. [DOI] [PubMed] [Google Scholar]
- 73.Knapp KG, Swartz JR. Cell-free production of active E. coli thioredoxin reductase and glutathione reductase. FEBS Lett. 2004;559(13):66–70. doi: 10.1016/S0014-5793(04)00025-0. [DOI] [PubMed] [Google Scholar]
- 74.Tewari YB, Goldberg RN. Thermodynamics of the oxidation–reduction reaction {2 glutathionered (aq)+ NADPox (aq)= glutathioneox (aq)+ NADPred (aq)} J Chem Thermodyn. 2003;35(8):1361–1381. [Google Scholar]
- 75.Bulock KG, Beardsley GP, Anderson KS. The kinetic mechanism of the human bifunctional enzyme atic (5-amino-4-imidazolecarboxamide ribonucleotide transformylase/inosine 5′-monophosphate cyclohydrolase). a surprising lack of substrate channeling. J Biol Chem. 2002;277(25):22168–22174. doi: 10.1074/jbc.M111964200. [DOI] [PubMed] [Google Scholar]
- 76.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36(Database issue):D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Willemos M, Hove-Jensen B. Binding of divalent magnesium by Escherichia coli phosphoribosyl diphosphate synthetase. Biochemistry. 1997;36(16):5078–5083. doi: 10.1021/bi962610a. [DOI] [PubMed] [Google Scholar]
- 78.Switzer RL. Regulation and mechanism of phosphoribosylpyrophosphate synthetase. i. purification and properties of the enzyme from Salmonella typhimurium. J Biol Chem. 1969;244(11):2854–2863. [PubMed] [Google Scholar]
- 79.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 80.Willsky GR, Malamy MH. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144(1):356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris RM, Webb DC, Howitt SM, Cox GB. Characterization of PitA and PitB from Escherichia coli. J Bacteriol. 2001;183(17):5008–5014. doi: 10.1128/JB.183.17.5008-5014.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amin N, Peterkofsky A. A dual mechanism for regulating cAMP levels in Escherichia coli. J Biol Chem. 1995;270(20):11803–11805. doi: 10.1074/jbc.270.20.11803. [DOI] [PubMed] [Google Scholar]
- 83.Traut T. Allosteric Regulatory Enzymes. Springer Verlag; 2007. [Google Scholar]
- 84.Rao NN, Torriani A. Molecular aspects of phosphate transport in Escherichia coli. Mol Microbiol. 1990;4(7):1083–1090. doi: 10.1111/j.1365-2958.1990.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 85.Alonso-Casajús N, Dauvillée D, Viale AM, Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Eydallin G, Ball S, Pozueta-Romero J. Glycogen phosphorylase, the product of the glgp gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J Bacteriol. 2006;188(14):5266–5272. doi: 10.1128/JB.01566-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the nonmevalonate pathway to terpenes: the role of the gcpe (ispg) protein. Proc Natl Acad Sci U S A. 2001;98(26):14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Komoto J, Yamada T, Takata Y, Markham GD, Takusagawa F. Crystal structure of the s-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of s-adenosylmethionine synthesis from atp and met. Biochemistry. 2004;43(7):1821–1831. doi: 10.1021/bi035611t. [DOI] [PubMed] [Google Scholar]
- 88.McQueney MS, Anderson KS, Markham GD. Energetics of s-adenosylmethionine synthetase catalysis. Biochemistry. 2000;39(15):4443–4454. doi: 10.1021/bi992876s. [DOI] [PubMed] [Google Scholar]
- 89.Schrimsher JL, Schendel FJ, Stubbe J, Smith JM. Purification and characterization of aminoimidazole ribonucleotide synthetase from Escherichia coli. Biochemistry. 1986;25(15):4366–4371. doi: 10.1021/bi00363a028. [DOI] [PubMed] [Google Scholar]
- 90.Rowe JJ, Lemmon RD, Tritz GJ. Nicotinic acid transport in Escherichia coli. Microbios. 1985;44(179180):169–184. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.