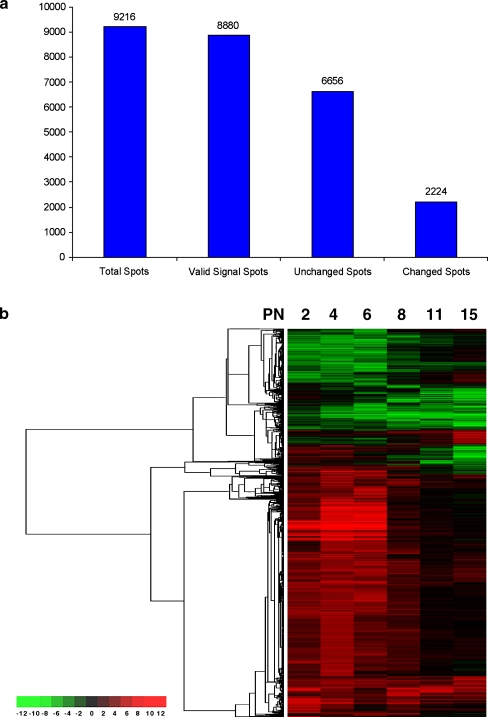
Fig. 2.
Overview of retinal gene expression in vitro and in vivo. a Numbers of changed and unchanged genes. The cDNA microarray used in this experiments contains 9,216 spots. 8,880 of them were valid signal spots (the spots which have the fluorescence intensity greater than 300 in any channel at any time point). 6,656 of 8,880 remained unchanged. 2,224 genes gained the mean ratio (in vitro vs in vivo) from two datasets greater than twofold (expression increased in in vitro at least twofold) and the ratio from each dataset greater than 1.5 or mean ratio less than 0.5 fold (expression decreased in in vitro at least twofold) and the ratio from each dataset less than 0.67. b Cluster analysis of the changed genes in all time points. Each numerical intensity ratio of in vitro/in vivo was assigned to a color on a scale from red to green, where red represents higher expression in vitro and green represents lower expression in vitro. There were about 25% genes changed. The first three time points contributed most of the changed genes. Many of them converged to a normal expression level later on. There was also a cluster of genes in the in vitro group, which started with a normal level but then decreased over time compared with in vivo. Many late development stage expression genes were in this group