Abstract
Animals regulate iron metabolism largely through the action of the iron regulatory proteins (IRPs). IRPs modulate mRNA utilization by binding to iron-responsive elements (IRE) in the 5′ or 3′ untranslated region of mRNAs encoding proteins involved in iron homeostasis or energy production. IRP1 is also the cytosolic isoform of aconitase. The activities of IRP1 are mutually exclusive and are modulated through the assembly/disassembly of its [4Fe–4S] cluster, reversibly converting it between an IRE-binding protein and cytosolic aconitase. IRP1 is also phosphoregulated by protein kinase C, but the mechanism by which phosphorylation posttranslationally increases IRE binding activity has not been fully defined. To investigate this, Ser-138 (S138), a PKC phosphorylation site, was mutated to phosphomimetic glutamate (S138E), aspartate (S138D), or nonphosphorylatable alanine (S138A). The S138E IRP1 mutant and, to a lesser extent, the S138D IRP1 mutant were impaired in aconitase function in yeast when grown aerobically but not when grown anaerobically. Purified wild-type and mutant IRP1s could be reconstituted to active aconitases anaerobically. However, when exposed to oxygen, the [4Fe–4S] cluster of the S138D and S138E mutants decayed 5-fold and 20-fold faster, respectively, than was observed for wild-type IRP1. Our findings suggest that stability of the Fe–S cluster of IRP1 can be regulated by phosphorylation and reveal a mechanism whereby the balance between the IRE binding and [4Fe–4S] forms of IRP1 can be modulated independently of cellular iron status. Furthermore, our results show that IRP1 can function as an oxygen-modulated posttranscriptional regulator of gene expression.
Iron is required by virtually all organisms and is utilized in a wide array of metabolic functions (1). In spite of its essential nature, iron can also be toxic, catalyzing the generation of damaging free radicals via the Fenton reaction (2). Consequently, organisms have evolved complex mechanisms for regulating iron transport, storage, and utilization (1, 3). In animals, iron metabolism is regulated largely through the action of iron regulatory proteins (IRPs), a small family of sequence-specific RNA binding proteins that regulate the utilization of mRNAs encoding proteins that function in the maintenance of iron homeostasis (4). IRPs bind to a conserved sequence called the iron-responsive element (IRE; refs. 4 and 5). IREs are located in either the 5′ or 3′ untranslated regions of mRNAs, where they mediate iron-responsive control of translation or mRNA stability, respectively. The role of IREs in modulating mRNA utilization has been characterized best for ferritin and transferrin receptor (TfR) mRNAs (4). IREs are also present in the mRNAs encoding erythroid-5-aminolevulinate synthase (6, 7), mitochondrial aconitase (7), succinate dehydrogenase subunit b of Drosophila melanogaster (8), and a divalent cation transporter, DCT1 (9). The diverse roles in iron metabolism of some of the proteins encoded by IRE-containing mRNAs indicates that IRPs are central components of a homeostatic network that permits the safe and efficient use of iron in animals.
Iron regulates the activity of IRP1 and IRP2 through fundamentally different mechanisms (4). High iron levels promote the assembly of a [4Fe–4S] cluster in IRP1, with concomitant loss of IRE binding activity (10, 11). Cluster assembly activates a second activity in IRP1, that of a cytosolic isoform of aconitase (c-acon; refs. 10 and 12). Regulation of IRP2 activity by iron is through protein degradation, with IRP2 being degraded in response to iron excess (13, 14). The consequence of these regulatory mechanisms is that IRP1 regulation by iron is reversible in the absence of protein synthesis, whereas IRP2 regulation is not.
The details of how cells regulate cluster assembly/disassembly in IRP1 remain undefined. It is clear that cellular iron status is a key component, but a number of observations have suggested that NO⋅ and oxygen and/or its metabolites also have an important role in modulating IRP1 function (15). Exposure of c-acon to oxidants results in conversion of the [4Fe–4S] cluster to a [3Fe–4S] cluster, and excess oxidant generates the apoprotein, which is the form of IRP1 that binds IRE (4, 10). NO⋅ and perhaps O2⨪ induce destruction of the Fe–S cluster of c-acon and enhance IRE binding activity in mammalian cells (15, 16). Administration of H2O2 to mammalian cells induces the IRE binding activity of IRP1 (15, 17), whereas hypoxia causes a decrease in this activity of IRP1, which is reversed by reoxygenation (18). Is there a role for cluster perturbants such as NO⋅ and/or O2⨪ in the regulation of IRP1 by iron? Furthermore, can the sensitivity of c-acon to such cluster perturbants be modulated? Elucidating the role of cluster stability in c-acon is central to our complete understanding of the means by which iron and other factors affect the function of IRP1.
Another mode of regulating IRP activity is through phosphorylation (19–21). Both IRP1 and IRP2 are phosphorylated by protein kinase C (PKC). IRP1 can be phosphorylated at two sites, Ser-138 (S138) and S711 (19, 20). Several pieces of evidence lead us to suggest that phosphorylation of IRP1 serves as a means to regulate its IRE binding activity and perhaps its interconversion with c-acon. First, treatment of HL60 cells with phorbol 12-myristate 13-acetate, a potent activator of PKC, stimulated phosphorylation of IRP1 and increased IRE binding activity and TfR mRNA abundance (19, 20). Second, the two PKC phosphorylation sites appear to be near the entrance to the proposed IRP1 active-site cleft (4, 19). Third, S138 is within a region of IRP1 required for high-affinity RNA binding (22, 23). Fourth, the RNA-binding form of IRP1 was a preferred substrate for phosphorylation, showing that PKC-dependent phosphorylation of IRP1 is directed toward the RNA binding form of the protein (21). These results suggest that phosphorylation is an iron-independent means of modulating the function of IRP1 through directed changes in the formation and/or function of the Fe–S cluster.
We provide evidence here that phosphorylation of IRP1 at S138 selectively affects its aconitase function by destabilizing the [4Fe–4S] cluster. Substitution of phosphomimetic amino acids for serine at position 138 causes enhanced susceptibility of the [4Fe–4S] cluster of IRP1 to attack by oxygen. We propose that phosphorylation of IRP1 at S138 renders its Fe–S cluster more susceptible to perturbants such as oxygen, H2O2, NO⋅, and O2⨪, or to other mechanisms that affect the kinetics of cluster assembly/disassembly, and therefore represents an iron-independent means of modulating iron metabolism. This mechanism of regulating IRP1 function illustrates its potential as an oxygen-modulated posttranscriptional regulator of gene expression.
MATERIALS AND METHODS
Strains and Culture Conditions.
Strain 0615d (a, ura3–52, trp1Δ63, his3Δ200, aco1, ade2) was used for all analysis of IRP1 aconitase function in vivo and was maintained on yeast extract/peptone/dextrose medium, or, when transformed with plasmids expressing IRP1, grown in synthetic complete medium lacking uracil and supplemented with 2% dextrose (SD-Ura; ref. 24). All yeast cultures were grown at 30°C. When yeast were grown anaerobically, the solid media were supplemented with 20 μg/ml ergosterol and 0.2% Tween 80 (25). The growth results shown are representative of results obtained with at least three transformants with each IRP1 mutant or wild-type (wt) gene.
Site-Directed Mutagenesis.
Mutagenesis of IRP1 was performed by the two-step PCR method (26). The pSP71 vector (Promega) was modified by insertion of a linker containing a DraIII site. The linker was inserted into the HindIII and XhoI sites of pSP71. A ClaI/DraIII fragment of the rabbit IRP1 cDNA (27) was cloned into the modified pSP71 vector. The first-step reactions (PCR I) were performed by using the SP6 primer along with one of the following mutagenic oligonucleotides that replaced Ser-138: Glu (TCTGTAATTCGTCAGCCCT), Asp (TCTGTAAATCGTCAGCCCT), or Ala (TCTGTAAAGCGTCAGCCCT). A 30-bp fragment containing the mutation of interest was then amplified by using PCR. PCR I reactions contained 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, 200 μM each dNTP, 2 μM each oligonucleotide, 1 ng of template DNA, and 2 units of Vent DNA polymerase (New England Biolabs) in a 50-μl final volume. PCR I reactions were amplified by using a 1-min denaturation at 94°C, a 2-min annealing at 40°C, a 2-min ramp to 72°C, and a 2-min of extension at 72°C. Forty cycles of amplification were performed. The 30-bp fragment produced in PCR I was used as a “megaprimer” for PCR reaction II. PCR II reactions consisted of 50 μl of PCR I plus 50 μl of reaction mixture similar to PCR I but containing 2 μM T7 primer and no additional template DNA. Amplification was performed as indicated for PCR I. After completion of the second round of amplification, the PCR II reactions were extracted with phenol/chloroform and precipitated with ethanol. A portion of the PCR II reaction was digested with ClaI and DraIII, purified by using agarose gel electrophoresis and Gene Clean (Bio 101), and subcloned into the ClaI and DraIII sites of the rabbit IRP1 cDNA. All mutagenesis reactions were confirmed by sequencing of the entire ClaI/DraIII fragment.
Enzyme and Protein Analysis.
Aconitase assays (1 ml) were performed by using 20 μl of yeast cytoplasmic extracts as described (28). Extracts were prepared from cells grown in SD-ura medium to mid-logarithmic phase. Cells were washed, resuspended in buffer [50 mM Tris⋅HCl, pH 8.0/50 mM KCl/2 mM citrate/10% glycerol/7 mM 2-mercaptoethanol/1 mM phenylmethylsulfonyl fluoride (PMSF)], and broken by repeated vortexing in the presence of 0.5-mm glass beads. Cell debris and unbroken cells were removed by centrifugation (16,000 × g for 5 min). Protein concentrations in cell extracts were determined spectrophotometrically (29).
His-tagged IRP1 was overexpressed in and purified from yeast as described (30). The yield of purified IRP1 was determined from amino acid analysis performed on each preparation. Reconstitution of the Fe–S cluster in purified IRP1 protein samples in the presence of Fe2+, DTT, cysteine, and NifS was performed as described (31). Enzyme activities were measured after anaerobic incubation at room temperature for ≥18 hr. The concentration of the resulting aconitase was calculated based on an activity of 3 units/nmol for the pure protein (28). EPR spectra of reconstituted IRP1 were recorded at 10–20 K on an X-band Varian E112 century series spectrometer as described in ref. 10. IRE binding analysis on purified IRP1 protein samples was performed by using internally labeled rat l-chain ferritin IRE as described (20).
RESULTS
The S138E Mutant of IRP1 Fails to Function Efficiently as an Aconitase in Aerobically Grown Yeast.
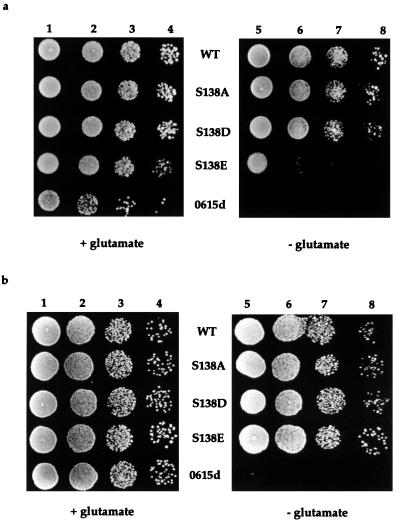
To investigate the effect of phosphorylation on IRP1 function, S138 was mutated to the phosphomimetic amino acids aspartate (S138D) or glutamate (S138E) or to nonphosphorylatable alanine (S138A), and the resulting IRP1s were analyzed for function after expression in aconitase-deficient (aco1) yeast, which are glutamate auxotrophs (32). We have found that IRP1 expressed from an ADH1 promoter can complement this trait in a dose-dependent manner, allowing them to grow in the absence of glutamate (R. Ma, J. Narahari, and W.E.W., unpublished observations). Expression of wt IRP1 permitted aco1 strain 0615d to grow nearly as well on media lacking glutamate as it did on glutamate-supplemented media (Fig. 1a). The S138A IRP1 mutant supported aconitase-dependent growth as well as wt IRP1 (Fig. 1a, WT and S138A in columns 7 and 8). Thus, the wt and S138A versions of IRP1 provided sufficient aconitase activity for glutamate biosynthesis in this strain. Yeast transformed with the S138D mutant grew well without glutamate but not as well as yeast expressing wt or S138A IRP1, suggesting that S138D mildly reduced aconitase activity in vivo (Fig. 1a). In contrast, yeast expressing the S138E IRP1 mutant grew very poorly on media lacking glutamate, indicating that the aconitase function of this mutant was significantly impaired in vivo under these growth conditions (Fig. 1a).
Figure 1.
Rescue of aco1 yeast from glutamate auxotrophy by expression of S138 IRP1 mutants. Yeast expressing either wild type (WT) or S138 (S138A, S138D, or S138E) IRP1 mutants were grown overnight in SD-Ura, washed twice with sterile H2O, and suspended into SD-Ura lacking glutamate. Nontransformed yeast (0615d) were grown in SD supplemented with uracil. Ten microliters containing between 2 × 105 and 2 × 102 yeast cells were spotted onto agar plates containing SD supplemented with uracil with or without glutamate as indicated. Growth after 4 days is shown. Columns 1 and 5, 2 × 105 cells; columns 2 and 6, 2 × 104 cells; columns 3 and 7, 2 × 103 cells; columns 4 and 8, 2 × 102 cells. (a) Aerobic growth. (b) Effect of anaerobiosis on growth of aco1 yeast expressing S138 IRP1 mutants on glutamate-free media. Growth analysis was performed as described above except that plates were incubated in an anaerobic chamber.
Enhancement of in Vivo Aconitase Function of the S138E IRP1 Mutant by Anaerobiosis.
Because the Fe–S cluster of aconitases is sensitive to oxygen (16, 33–35), we investigated whether anaerobiosis influenced the aconitase activity of the wt and mutant versions of IRP1 expressed in aco1 yeast. Yeast expressing the wt IRP1 or the S138A mutant grew essentially the same with or without glutamate irrespective of the presence or absence of oxygen (compare Fig. 1 a and b). Yeast expressing the S138D mutant IRP1 also grew vigorously in anaerobic conditions in the absence of glutamate. Most strikingly, growth of yeast expressing the S138E mutant was substantially enhanced by anaerobiosis to a level equivalent to that seen for the wt IRP1-expressing yeast (Fig. 1b). The inability of nontransformed yeast to grow on media lacking glutamate was not reversed by anaerobiosis, indicating that the effect depended on IRP1 aconitase function (Fig. 1). Collectively, our results suggest that oxygen and/or its metabolites causes inactivation of the aconitase function of the S138E IRP1 mutant in cells growing aerobically, perhaps by causing disassembly of the [4Fe–4S] cluster in vivo.
Absence of Aconitase Activity in Aerobically Prepared Cytoplasmic Extracts of Yeast Expressing Phosphomimetic IRP1 Mutants.
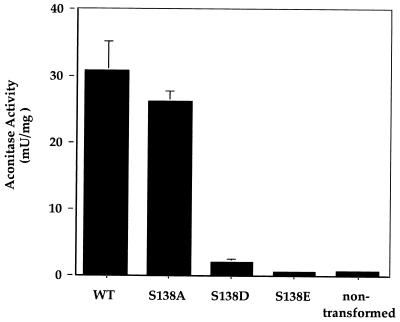
We determined the aconitase activity in aerobically prepared extracts from aco1 yeast expressing wt or S138 mutant versions of IRP1 to examine the relationship between aconitase activity in vitro and cell growth in media lacking glutamate. Extracts of nontransformed yeast gave a background activity of 0.8 milliunits/mg (Fig. 2). Aconitase activity nearly 40-fold above background (30.8 ± 4.4 milliunits/mg) was detected in extracts of yeast transformed with wt IRP1. A similar level of aconitase activity was observed in extracts of yeast expressing the S138A mutant (26.1 ± 1.7 milliunits/mg). The relative activity found for the S138A mutant and wt IRP1 is consistent with the ability of these IRP1s to support similar growth rates in glutamate-free media (Fig. 1). In contrast, extracts from yeast expressing the S138D mutant displayed aconitase activity (2.1 ± 0.5 milliunits/mg) that was only 7% of that found in extracts of yeast expressing wt IRP1, whereas extracts from yeast expressing the S138E mutant had aconitase activity no different than background (0.6 ± 0.3 milliunits/mg; Fig. 2). Note that all IRP1s were expressed to a similar level in the yeast (results not shown). Interestingly, reduction in measurable aconitase activity from wt IRP1 to a level comparable to that measured in vitro for the S138D IRP1 mutant (obtained by lowering wt IRP1 expression in vivo) significantly impairs the growth of this aco1 strain in glutamate-free media (J. Narahari and W.E.W., unpublished results), whereas this strain expressing the S138D mutant IRP1 grew nearly as well as when expressing wt IRP1 (Fig. 1). In addition, extracts of yeast expressing the S138E mutant exhibited no aconitase activity over background levels, but yeast expressing this IRP1 mutant exhibited limited aerobic growth and vigorous anaerobic growth on glutamate-free media. In contrast, the nontransformed aco1 yeast failed to grow without glutamate supplementation under any condition (Fig. 1). We conclude that both the S138D and the S138E IRP1 mutants were active aconitases in the intact yeast cell, but had lost most of this activity during extract preparation.
Figure 2.
Aconitase activity of S138 IRP1 mutants in aerobically prepared cytoplasmic extracts of aco1 yeast. Yeast expressing the indicated IRP1 mutants were grown to mid-logarithmic phase, harvested, and cytoplasmic extracts were prepared and analyzed for aconitase activity. Activities shown are per mg of total extracted protein and are an average of three independent experiments. Error bars indicate SD.
Activation of Aconitase Function in Phosphomimetic IRP1 Mutants upon Anaerobic Reconstitution of the Fe–S Cluster.
To gain further insight into the effect of mutation of S138 on the aconitase function of IRP1, we reconstituted the Fe–S cluster in purified wt or S138 mutant versions of IRP1 anaerobically and determined their aconitase activity. wt IRP1 yielded an average specific activity of 19.8 ± 3.5 units/mg protein after reconstitution (Table 1). A similar level of activity was reconstituted in the S138A and S138D mutant IRP1s, and substantial aconitase activity was reconstituted in the S138E IRP1, although it was routinely about 40% lower than wt IRP1 (Table 1). These results demonstrate that IRP1 containing phosphomimetic substitutions at residue 138 could function as an aconitase upon reconstitution of the [4Fe–4S] cluster under anaerobic conditions in vitro. Although the S138E mutation did result in a reduction in the aconitase activity measured for the mutant IRP1 after reconstitution, it is important to note that this mutation did not ablate IRP1’s aconitase function. Therefore, the differences in relative aconitase activity measured in cytoplasmic extracts of aco1 yeast expressing these mutant IRP1s (Fig. 2) was caused by differences in the proportion of the IRP1s that had a [4Fe–4S] cluster. The reason for this is likely a difference in cluster stability during extract preparation, but could also reflect differences in cluster stability and/or assembly in the intact cell.
Table 1.
Aconitase and IRE binding functions of purified S138 IRP1 mutants
| IRP1 | Reconstituted aconitase activity, units/mg | IRE binding affinity of apo-IRP1, pM |
|---|---|---|
| wt | 19.8 ± 3.5 | 61 ± 19 |
| S138A | 24.3 ± 1.5 | 23 ± 6.3 |
| S138D | 18.5 ± 1.1 | 36 ± 18.5 |
| S138E | 10.8 ± 1.8 | 19 ± 7 |
The various IRP1 mutants were purified from yeast and subjected to anaerobic Fe–S cluster reconstitution followed by aconitase assay, or the apoprotein was analyzed for IRE-binding ability. Reconstitution and analysis of aconitase activity was performed on a minimum of three independent preparations of each mutant IRP1. Saturation curves to determine the affinity of IRP1–IRE interaction were done a minimum of four times representing at least two independent experiments for each mutant IRP1. Values shown are ± SD.
Instability of the Fe–S Cluster of the S138D and S138E IRP1 Mutants.
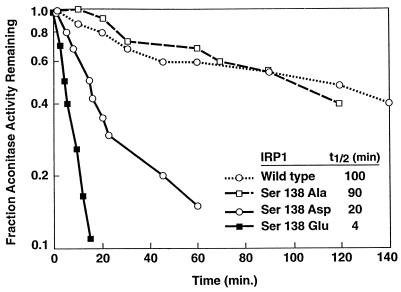
The characteristics of the IRP1 mutants containing phosphomimetic substitutions for S138 suggest that the [4Fe–4S] cluster of these proteins is more sensitive to oxygen and other cluster perturbants, than is the wt protein. To investigate this, purified IRP1 mutants were reconstituted anaerobically and exposed to air-saturated buffer as a source of oxygen. Enzyme activity was monitored over time as a measure of the presence of the [4Fe–4S] cluster. Wild-type IRP1 and the S138A mutant were similarly stable to this treatment, displaying a half-life for enzyme activity of ≈100 min and 90 min, respectively (Fig. 3). The aconitase activity of the S138D mutant decayed 5-fold faster than wt IRP1. Even more striking was the S138E mutant, whose aconitase activity decayed with a half-life of 4 min, or more than 20-fold faster than wt IRP1 (Fig. 3). These results demonstrate that the stability in the presence of oxygen of the [4Fe–4S] cluster of IRP1 mutants containing phosphomimetic substitutions for S138 was very much reduced and are consistent with the view that phosphorylation of S138 decreases the stability of the [4Fe–4S] cluster of IRP1 in vivo.
Figure 3.
Half-life of aconitase activity of S138 IRP1 mutants upon exposure to oxygen. IRP1 mutants were overexpressed and purified from yeast and subjected to in vitro cluster reconstitution anaerobically. Aconitase activity was measured at various times after diluting the reconstituted protein into air-saturated buffer. The half-life (t1/2) of aconitase activity for each IRP1 mutant is indicated in the inset.
To examine the effect of oxidation on the Fe–S cluster in the wt and mutant IRP1s, each was reconstituted anaerobically, exposed to air-saturated buffer, and analyzed by EPR spectroscopy. Oxidation of aconitases results in conversion of the [4Fe–4S] cluster to a [3Fe–4S] cluster, and excess oxidant can cause complete cluster removal (16, 33–35). Typical spectra for the [3Fe–4S] cluster of c-acon were obtained for both the wt and S138A IRP1s upon air oxidation (ref. 10; results not shown). Similar analysis of the S138D mutant also yielded spectra for the [3Fe–4S] cluster. However, the intensity of the signal obtained was less than 20% that observed for wt IRP1 when corrections for differences in protein concentration were made. No [3Fe–4S] spectra were obtained for the S138E mutant. Similar results were obtained with ferricyanide oxidation of the reconstituted IRP1s (results not shown). The reduction in the intensity of the EPR signal for the S138D mutant and the absence of a signal for the S138E mutant indicate that there had been significant loss of both the [4Fe–4S] and the [3Fe–4S] forms of the Fe–S cluster from the phosphomimetic mutants of IRP1 upon oxygen or ferricyanide oxidation. In contrast, much less cluster loss occurred from the wt or S138A IRP1s. These results lend support to the view that IRP1 has the capacity to serve as a biosensor of oxygen.
Lack of an Effect on IRE Binding of Mutations at S138 of IRP1.
To determine whether mutation of S138 had a direct effect on IRE binding, quantitative IRE-binding analysis was performed. The wt and mutant IRP1s were overexpressed in and purified from yeast, and the pure apoprotein was analyzed for IRE-binding affinity. Wild-type IRP1 bound a rat l-ferritin IRE with a KD of 61 ± 19 pM (Table 1). This was within the range of the affinities previously reported for native IRP1 (36–38). The binding affinity obtained for each of the IRP1 mutants was in the range of the affinity for native IRP1 for this IRE as well (Table 1). Thus, these mutations of S138 do not negatively affect the IRP1–IRE interaction. This is similar to the effect of inducing phosphorylation of IRP1 by PKC in HL60 cells, where total IRE binding is increased but its affinity is unaffected (20).
DISCUSSION
The readiness with which cluster assembly/disassembly occurs in IRP1 in vivo makes it unusual among iron–sulfur proteins. Cluster disassembly occurs under iron-limiting conditions in animal cells, although the molecular mechanism by which this occurs is not at all understood. The data presented here suggest a mechanism for regulating Fe–S cluster turnover and the interconversion of IRP1 and c-acon through phosphorylation of S138. Our results show that substitution of either Asp or Glu, which can mimic the charge and/or size effect of phosphorylation, for S138 results in a marked instability of the Fe–S cluster in c-acon, particularly in the presence of oxygen. It is well established that the [4Fe–4S] cluster in aconitases are particularly sensitive to oxygen and other perturbants (16, 33–35). In fact, results from other studies have suggested a role for reactive oxygen intermediates in the disassembly of the [4Fe–4S] cluster of IRP1 (4, 15). In the model proposed here, phosphorylation of IRP1 at S138 greatly enhances the rate of Fe–S cluster disassembly, thus favoring the accumulation of the RNA-binding form of IRP1. This accumulation would lead to an increase in total IRE binding activity in the absence of a change in iron availability, and thus is an iron-independent mode of regulating IRP1 function. Physiologically, this regulation could be important when cells need to alter the uptake and/or metabolic fate of iron. This situation could occur during monocytic or erythropoietic differentiation and cell proliferation. These circumstances require increased levels of cellular iron concomitant with an increase in IRE binding activity needed for the accumulation of TfR mRNA (39–43). The ability to differentially regulate IRP activity independently of cellular iron status is crucial to the normal progression of such processes. A tissue-specific pattern of IRP1 phosphorylation in conjunction with differential expression of IRP1 and IRP2 could have profound consequences for organismal iron homeostasis. To our knowledge, such an effect of phosphorylation on the stability of the Fe–S cluster in IRP1 where phosphorylation regulates the stability of a metal cluster has not been observed previously.
Clearly, our results also point to IRP1 serving a role as a biosensor of oxygen. Other investigators have proposed that IRP1 could be regulated by oxidative stress through an indirect mechanism (15, 18). An important distinction with our model is that we propose that IRP1 is a direct target for oxygen-mediated cluster disassembly. This is analogous to the prokaryotic transcription factors FNR and SoxR, where oxygen and O2⨪, respectively, regulate the activity of these proteins through a direct effect on their Fe–S cluster (44–47). For IRP1, the phosphorylated protein would have an enhanced responsiveness to physiological and pathological fluctuations in oxygen and/or oxygen metabolites, providing an additional level of regulation by kinases. Such a regulatory mechanism could have important consequences in certain pathophysiological processes such as in cells of the reticuloendothelial system during the inflammatory response and in ischemia reperfusion injury, where iron in the presence of oxygen is thought to be cytotoxic (18, 48). Finally, a provocative notion is that IREs exist in the mRNAs of oxygen-regulated genes and that these mRNAs would be preferentially regulated by IRP1.
Aconitase-deficient yeast can use the aconitase activity of IRP1 to generate precursors for glutamate biosynthesis and thus are able to grow on glutamate-free media with IRP1 protein as the sole source of aconitase activity. Phosphomimetic amino acid substitution for S138 of IRP1 makes the [4Fe–4S] cluster in c-acon unstable such that the normal production of cluster perturbants, such as superoxides (49, 50), causes disruption of the [4Fe–4S] cluster. This impairs aconitase function in vivo and limits growth of aco1 yeast expressing these mutant IRP1s in the absence of glutamate. This phenotype was most severe when glutamate was substituted for S138, though aspartate substitution caused enhanced oxygen-mediated decay of aconitase activity as well. Several other aco1 yeast strains showed growth defects on glutamate-free media when transformed with the S138D IRP1 mutant, indicating that the instability of the Fe–S cluster of this mutant could have strong detrimental effects in vivo as well (results not shown). The detrimental effect of the S138E mutation on the aconitase function of IRP1 could be suppressed in vivo by growth of yeast under anaerobic conditions. This effect of anaerobiosis on the aco1 yeast expressing the S138E IRP1 mutant is reminiscent of sod1 yeast, which display several amino acid auxotrophies when grown aerobically that can be suppressed by anaerobic growth (50, 51). The auxotrophies of sod1 yeast are thought to result from superoxide-mediated destruction of Fe–S clusters in key enzymes of the affected amino acid biosynthetic pathways (49, 52, 53). These observations raise the question as to whether normal production of reactive oxygen intermediates and other cluster perturbants in animal cells is an important component in the regulation of IRP1 function and therefore iron metabolism. If such is the case, then defects in oxygen regulation, for instance in amyotrophic lateral sclerosis (49, 54), would not only lead to an increase of potentially toxic reactive oxygen intermediates but would also alter iron regulation, which could exacerbate the condition.
The molecular basis for the instability effects of Glu and Asp substitution for S138 on the Fe–S cluster of IRP1 is not clear. When modeled on the mitochondrial aconitase structure, S138 of IRP1 is not predicted to be near the Fe–S cluster, nor would it be expected to lie within the enzyme active site (55, 56). S138 is predicted to lie near the entrance to the active-site cleft, however (4, 19, 56). This then suggests that phosphorylation of S138 may affect Fe–S cluster stability indirectly, perhaps by distorting the active-site cleft. The fact that aconitase activity can be reconstituted under anaerobic conditions in vitro in all of the mutant IRP1s argues that this distortion is not sufficient to ablate enzymatic activity. For the S138E mutant, there was a reduction in the amount of activity that could be reconstituted in vitro. However, at this time we cannot distinguish between an effect of this mutation on enzymatic function or strictly on Fe–S cluster stability. The very rapid decay of the aconitase activity in this mutant upon exposure to oxygen and the enhancement of in vivo activity under anaerobic conditions suggests that the major effect of this mutation is on [4Fe–4S] cluster stability.
It is likely that phosphorylation at S138 causes only a local conformational change in IRP1, which is incompatible with maintaining a stable Fe–S cluster. This is suggested by the observation that neither Glu nor Asp substitution for S138 had an effect on IRE binding. It has been proposed that IRP1 can adopt one of two conformations, one allowing high-affinity IRE binding and the second allowing accommodation of an Fe–S cluster (21). Assembly of an Fe–S cluster converts the protein to the aconitase form and inhibits IRE binding. An interesting possibility is that phosphorylation of S138 favors the IRE binding conformation of IRP1, which is incompatible with a stable cluster and the aconitase form. This would suggest that S138 resides in a region of the protein in which a conformational change accompanies the assembly/disassembly of the Fe–S cluster. Although there is no direct evidence for this, Schalinske et al. (21) have observed that reactivity of a major chymotrypsin cleavage site adjacent to S138 is substantially reduced by the presence of an Fe–S cluster in IRP1.
The fact that none of the substitutions for S138 had an effect on binding to a ferritin IRE was surprising because S138 lies within a region of IRP1 that is in contact with the IRE when bound (22, 23). This suggests that S138 is not itself a critical residue for IRE binding and that its side chain most likely points away from the IRE binding pocket. This view is supported by the finding that the increased size of the side chains of Glu and Asp in comparison to Ser at position 138 had no effect on IRE binding. On the other hand, S138 could reside within a flexible region on IRP1 such that a variety of amino acids can be accommodated for IRE binding. In either case, S138 does not itself appear to be an important residue for IRE recognition or binding affinity in IRP1 even though it has a profound effect on the stability of its Fe–S cluster.
Regulation of IRP1 by iron is a well established phenomenon. This study provides insights into a mechanism for the control of IRP1 function and suggests that PKC and/or other kinases that phosphorylate S138 may also play a role in iron homeostasis through their effects on IRP1. Previous studies have implicated phosphorylation of IRPs as a mode of inducing IRE binding activity (18–20, 57). Our observations illustrate how phosphorylation could cause this effect by affecting Fe–S cluster stability. The effect of anaerobiosis on the activity of the S138E mutant IRP1 in vivo suggests that constitutive and inducible production of reactive oxygen intermediates normally plays a pivotal role in the disassembly of the Fe–S cluster in IRP1 in animal cells.
Acknowledgments
Supported by grants from the National Institutes of Health to W.E.W. (DK-47281), R.S.E. (DK-47219), and M.C.K. (GM-51831) and from the University of Wisconsin College of Agriculture and Life Sciences Hatch Project 3951 (R.S.E.). We acknowledge the use of the facilities of the National Biomedical ESR Center at the Medical College of Wisconsin (supported by National Institutes of Health Grant RR-01008). T.B.C. was supported by a U.S. Department of Agriculture National Needs Fellowship.
ABBREVIATIONS
- IRP
iron regulatory protein
- IRE
iron-responsive element
- c-acon
cytoplasmic aconitase
- PKC
protein kinase C
- S138
Ser-138
- TfR
transferrin receptor
References
- 1.Crichton R R. Inorganic Biochemistry of Iron Metabolism. New York: Ellis Horwood; 1991. [Google Scholar]
- 2.Halliwell B, Gutteridge J M C. Arch Biochem Biophys. 1986;246:501–514. doi: 10.1016/0003-9861(86)90305-x. [DOI] [PubMed] [Google Scholar]
- 3.Williams R J P. In: Iron Transport and Storage. Ponka P, Schulman H M, Woodworth R C, editors. Boca Raton, FL: CRC; 1990. pp. 1–16. [Google Scholar]
- 4.Eisenstein R S, Kennedy M C, Beinert H. In: Metal Ions in Gene Regulation. Silver S, Walden W, editors. New York: International Thomson; 1997. pp. 157–216. [Google Scholar]
- 5.Hentze M W, Caughman S W, Rouault T A, Barriocanal J G, Dancis A, Harford J B, Klausner R D. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 6.Cox T C, Bawden M J, Martin A, May B K. EMBO J. 1991;10:1891–1902. doi: 10.1002/j.1460-2075.1991.tb07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandekar T, Stripecke R, Gray N K, Goossen B, Constable A, Johansson H E, Hentze M W. EMBO J. 1991;10:1903–1909. doi: 10.1002/j.1460-2075.1991.tb07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler S A, Henderson B R, Kühn L C. J Biol Chem. 1995;270:30781–30786. doi: 10.1074/jbc.270.51.30781. [DOI] [PubMed] [Google Scholar]
- 9.Gunshin H, Mackenzie B, Berger U, Gunshin Y, Romero M, Boron W, Nussberger S, Gollan J, Hediger M. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy M C, Mende-Mueller L, Blondin G A, Beinert H. Proc Natl Acad Sci USA. 1992;89:11730–11734. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haile D J, Rouault T A, Harford J B, Kennedy M C, Blondin G A, Beinert H, Klausner R D. Proc Natl Acad Sci USA. 1992;89:11735–11739. doi: 10.1073/pnas.89.24.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaptain S, Downey W E, Tang C, Philpott C, Haile D, Orloff D G, Harford J B, Rouault T A, Klausner R D. Proc Natl Acad Sci USA. 1991;88:10109–10113. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo B, Phillips J D, Yu Y, Leibold E A. J Biol Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 14.Samaniego F, Chin J, Iwai K, Rouault T A, Klausner R D. J Biol Chem. 1994;269:30904–30910. [PubMed] [Google Scholar]
- 15.Hentze M W, Kühn L C. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy M C, Antholine W E, Beinert H. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 17.Cairo G, Pietrangelo A. Eur J Biochem. 1995;232:358–363. doi: 10.1111/j.1432-1033.1995.358zz.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanson E, Leibold E A. J Biol Chem. 1998;273:7588–7593. doi: 10.1074/jbc.273.13.7588. [DOI] [PubMed] [Google Scholar]
- 19.Eisenstein R S, Tuazon P T, Schalinske K L, Anderson S A, Traugh J A. J Biol Chem. 1993;268:27363–27370. [PubMed] [Google Scholar]
- 20.Schalinske K L, Eisenstein R S. J Biol Chem. 1996;271:7168–7176. doi: 10.1074/jbc.271.12.7168. [DOI] [PubMed] [Google Scholar]
- 21.Schalinske K L, Anderson S A, Tuazon P T, Chen O S, Kennedy M C, Eisenstein R S. Biochemistry. 1997;36:3950–3958. doi: 10.1021/bi9624447. [DOI] [PubMed] [Google Scholar]
- 22.Basilion J P, Rouault T A, Massinople C M, Klausner R D, Burgess W H. Proc Natl Acad Sci USA. 1994;91:574–578. doi: 10.1073/pnas.91.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neupert B, Menotti E, Kühn L C. Nucleic Acids Res. 1995;23:2579–2583. doi: 10.1093/nar/23.14.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 25.Hassett R F, Romeo A M, Kosman D J. J Biol Chem. 1998;273:7628–7636. doi: 10.1074/jbc.273.13.7628. [DOI] [PubMed] [Google Scholar]
- 26.Marini F, Naeem A, Lapeyre J N. Nucleic Acids Res. 1993;21:2277–2278. doi: 10.1093/nar/21.9.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patiño M M, Walden W E. J Biol Chem. 1992;267:19011–19016. [PubMed] [Google Scholar]
- 28.Kennedy M C, Emptage M H, Dreyer J L, Beinert H. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 29.Groves W E, Davis F C, Sells B H. Anal Biochem. 1968;22:195–210. doi: 10.1016/0003-2697(68)90307-2. [DOI] [PubMed] [Google Scholar]
- 30.Ke Y, Wu J, Leibold E A, Walden W E, Theil E C. J Biol Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- 31.Basilion J P, Kennedy M C, Beinert H, Massinople C M, Klausner R D, Rouault T A. Arch Biochem Biophys. 1994;311:517–522. doi: 10.1006/abbi.1994.1270. [DOI] [PubMed] [Google Scholar]
- 32.Ogur M, Coker L, Ogur S. Biochem Biophys Res Commun. 1964;14:193–197. doi: 10.1016/0006-291x(64)90254-2. [DOI] [PubMed] [Google Scholar]
- 33.Gardner P R, Fridovich I. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 34.Gardner P R, Raineri I, Epstein L B, White C W. J Biol Chem. 1995;270:13399–13405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 35.Kent T A, Dreyer J L, Kennedy M C, Huynh B H, Emptage M H, Beinert H, Münck E. Proc Natl Acad Sci USA. 1982;79:1096–1100. doi: 10.1073/pnas.79.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barton H A, Eisenstein R S, Bomford A, Munro H N. J Biol Chem. 1990;265:7000–7008. [PubMed] [Google Scholar]
- 37.Haile D J, Hentze M W, Rouault T A, Harford J B, Klausner R D. Mol Cell Biol. 1989;9:5055–5061. doi: 10.1128/mcb.9.11.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson G R, Patiño M M, Beck M M, Gaffield L, Walden W E. Biol Met. 1991;4:48–55. doi: 10.1007/BF01135557. [DOI] [PubMed] [Google Scholar]
- 39.Testa U, Petrini M, Quaranta M T, Pelosi-Testa E, Mastroberardino G, Camagna A, Boccoli G, Sargiacomo M, Isacchi G, Cozzi A, et al. J Biol Chem. 1989;264:13181–13197. [PubMed] [Google Scholar]
- 40.Testa U, Kühn L, Petrini M, Quaranta M T, Pelosi E, Peschle C. J Biol Chem. 1991;266:13925–13930. [PubMed] [Google Scholar]
- 41.Andreesen R, Osterholz J, Bodemann H, Bross K J, Costabel U, Lohr G W. Blut. 1984;49:195–202. doi: 10.1007/BF00319822. [DOI] [PubMed] [Google Scholar]
- 42.Seiser C, Teixeira S, Kühn L C. J Biol Chem. 1993;268:13074–13080. [PubMed] [Google Scholar]
- 43.Teixeira S, Kühn L C. Eur J Biochem. 1991;202:819–826. doi: 10.1111/j.1432-1033.1991.tb16438.x. [DOI] [PubMed] [Google Scholar]
- 44.Demple B. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 45.Unden G, Schirawski J. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 46.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley P. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidalgo E, Ding H, Demple B. Cell. 1997;88:121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 48.Recalcati S, Taramelli D, Conte D, Cairo G. Blood. 1998;91:1059–1066. [PubMed] [Google Scholar]
- 49.Fridovich I. J Biol Chem. 1997;272:18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 50.Moradas-Ferreira P, Costa V, Piper P, Mager W. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 51.Bilinski T, Krawiec Z, Liczmanski A, Litwinska J. Biochem Biophys Res Commun. 1985;130:533–539. doi: 10.1016/0006-291x(85)90449-8. [DOI] [PubMed] [Google Scholar]
- 52.Flint D H, Tuminello J F, Emptage M H. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 53.Flint D H, Smyk-Randall E, Tuminello J F, Draczynska-Lusiak B, Brown O R. J Biol Chem. 1993;268:25547–25552. [PubMed] [Google Scholar]
- 54.Brown R H., Jr Arch Neurol. 1997;54:1246–1250. doi: 10.1001/archneur.1997.00550220050013. [DOI] [PubMed] [Google Scholar]
- 55.Lauble H, Kennedy M C, Beinert H, Stout C D. Biochemistry. 1992;31:2735–2748. doi: 10.1021/bi00125a014. [DOI] [PubMed] [Google Scholar]
- 56.Klausner R D, Stout C D, Rouault T A. Chem Biol. 1994;1:viv–xv. [Google Scholar]
- 57.Pantopoulos K, Hentze M W. EMBO J. 1995;14:2917–2924. doi: 10.1002/j.1460-2075.1995.tb07291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]