Abstract
The 46 kDa cation-dependent MPR (CD-MPR) plays a key role in the delivery of lysosomal enzymes to the lysosome by binding newly synthesized mannose 6-phosphate (Man-6-P)-containing acid hydrolases and diverting them from the secretory pathway. Previous studies on a truncated form of the receptor comprised of only the soluble extracellular region (sCD-MPR, residues 1-154) have shown that the CD-MPR exists as a homodimer and exhibits two distinct conformations in the ligand-bound versus ligand-unbound states, involving changes in quaternary structure and positioning of loop D, the residues of which form a side of the binding pocket in the presence of ligand. To determine the role of inter-monomer contacts in the functioning of the sCD-MPR, site-directed mutagenesis was used to generate a construct lacking a salt bridge (Glu19-Lys137) that tethers the N-terminal α-helix of one subunit to loop D of the other subunit in the ligand-bound form. Here we show by surface plasmon resonance analyses and NMR spectroscopy that the elimination of this inter-monomer salt bridge significantly decreases binding affinity of the mutant receptor (E19Q/K137M) towards lysosomal enzymes and Man-6-P. Analyses of the E19Q/K137M mutant receptor crystallized under various conditions revealed an altered quaternary structure that is intermediate between those observed in the ligand-bound and ligand-unbound states. Taken together, the results demonstrate a key role for inter-monomer interactions in the structure and functioning of the CD-MPR.
The biogenesis of lysosomes requires the correct sorting of >60 acid hydrolases from their site of synthesis in the endoplasmic reticulum to their final destination in lysosomes (1). This targeting event is mediated by the 46 kDa cation-dependent mannose 6-phosphate receptor (CD-MPR) and the 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR), the only two members of the P-type family of lectins (2–4). The two MPRs recognize phosphomannosyl residues on high mannose-type N-glycans that serve to mark newly synthesized soluble acid hydrolases for delivery to endosomal/lysosomal compartments. The MPRs transport their cargo from the trans Golgi network (TGN) to endosomes where the acidic pH of the endosomal compartment facilitates disassembly of the complex. The released lysosomal enzymes are packaged into lysosomes while the receptors either return to the Golgi/TGN via a retromer-assisted transport system (5) to repeat the process or move to the plasma membrane where the CI-MPR, but not the CD-MPR, functions to internalize exogenous ligands at the cell surface (6, 7). Consistent with its role in intracellular trafficking, biochemical studies have confirmed that the CD-MPR displays strict pH dependence with respect to ligand binding, with optimal binding occurring at the pH of the TGN (~pH 6.5) and little or no binding occurring at pH values above pH 7.4 or below pH 5.5, corresponding to values found at the cell surface and endosomes, respectively (7, 8). Both the CI-MPR and CD-MPR are required to transport the entire repertoire of lysosomal enzymes to lysosomes, and a recent report indicates that the CD-MPR transports several lysosomal enzymes not recognized by the CI-MPR (9).
The CD-MPR is a type I membrane glycoprotein that exists as a homodimer, with each subunit containing a single Man-6-P binding site (8). We previously reported the crystal structure of the extracellular region comprising residues 1-154 of the CD-MPR (sCD-MPR) in the absence of ligand at pH 6.5 (10) which shows that the ligand-free form of the extracytoplasmic region of the CD-MPR crystallized as a dimer and maintained a similar overall topology as the ligand-bound form at pH 6.5 (11). Surprisingly, unlike other lectins which do not display any significant conformational change induced by ligand binding, a dramatic change in quaternary structure was revealed: the CD-MPR monomers undergo significant movements relative to each other, resulting in an open conformation in all structures containing ligand in the binding pocket, and a closed conformation found in all structures lacking bound carbohydrate (10, 12). Accompanying this change in quaternary structure, one (i.e., loop D) of the two essential loops that comprise the binding pocket repositions itself in the absence of ligand to a location held by the carbohydrate ligand. Thus, the integrity of the binding site is preserved as the side chains of three (Gln66, Arg111 and Tyr143) out of the four amino acids essential for recognition of the mannose ring maintain their ligand binding position due to this reorientation of loop D (10, 12).
A comparison of the ligand-free and ligand-bound crystal structures of the CD-MPR revealed a solvent accessible inter-monomer salt bridge, Glu19 from the N-terminal α-helix of one monomer and Lys137 from loop D of the other monomer, present only in the ligand-bound form. It was hypothesized that disruption of this inter-monomer salt bridge could contribute to the pH sensitive release mechanism of the CD-MPR (12). In the current report, the contributions of the inter-monomer salt bridge between Glu19 and Lys137 to the functioning of the CD-MPR were evaluated both biochemically and biophysically. A CD-MPR mutant (E19Q/K137M) containing two amino acid substitutions (Glu19 was replaced with Gln, and Lys137 was replaced with Met) was generated and analyzed for its ability to bind: 1) lysosomal enzymes by quantitative surface plasmon resonance measurements, and 2) the monosaccharide, Man-6-P by NMR spectroscopy. Furthermore, the crystal structure of E19Q/K137M was determined under different conditions. Taken together, the results demonstrate the critical role this ionic interaction plays in stabilizing a high affinity binding pocket and further highlights a mechanistic rationale for how this receptor functions as a dimer to binding lysosomal enzymes. Furthermore, the results show that the CD-MPR is dynamic, and suggests that the E19Q/K137M mutant structure represents an intermediate along the pathway of cargo loading and unloading.
EXPERIMENTAL PROCEDURES
Materials
The following reagents were obtained commercially as indicated: restriction endonucleases (New England Biolabs); Pichia pastoris wild-type strain X-33, P. pastoris expression vectors pGAPZαA and pPICZαA, T4 DNA ligase, BenchMark prestained protein ladder, and Zeocin (Invitrogen Corp.); glucose 6-phosphate, Man-6-P, Concanavalin A (ConA) Sepharose (Sigma); protein A-horseradish peroxidase conjugate (Amersham Biosciences); Immobilon P (Millipore); SuperSignal West Pico chemiluminescent substrate (Pierce Chemical Corp.); nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen); amine coupling kits, surfactant P20 and CM5 research grade sensor chips (GE Healthcare). Detergent screening kit (Hampton Research); MTX 3.2 cells that over-express human β-glucuronidase were generously provided by Dr. W. Sly (St. Louis University School of Medicine, St. Louis, MO). Phosphomannan from Hansenula holstii was a kind gift of Dr. M. E. Slodki (Northern Regional Research Center, Peoria, IL).
Generation of Constructs
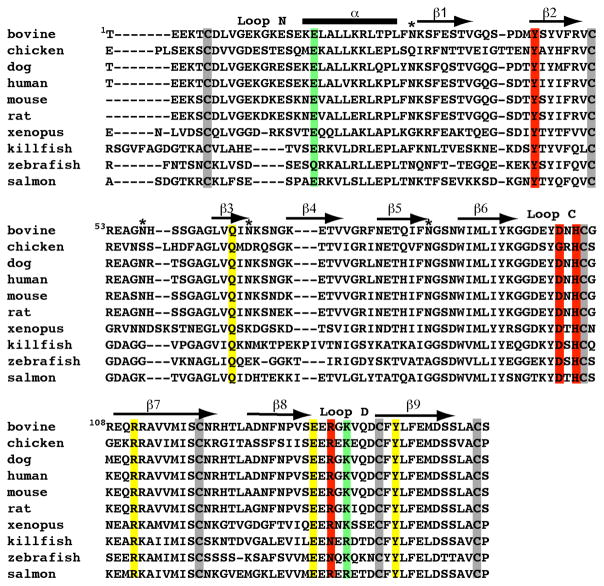
Plasmids containing a glycosylation-deficient form of the extracytoplasmic domain (residues 1-154) of the bovine CD-MPR in which asparagine residues at positions 31, 57, 68, and 87 were replaced with glutamine (Fig. 1) to eliminate four of the five potential N-linked glycosylation sites, leaving the N-glycosylation site at position 81 intact, with (sCD-MPRHis6) (13) or without (sCD-MPR) (14) a C-terminal tag composed of six histidine residues were modified to contain two additional amino acid substitutions to eliminate the salt bridge located between the two subunits of the dimer. E19Q and K137M substitutions were generated by oligonucleotide-directed mutagenesis according to the method of Kunkel et al. as described previously (15). The mutagenic oligonucleotides used are: E19Q (5′-TTT GTT GAT CTC CAC CAG GC-3′), K137M (5′-ACA GTG GTT CTC ATA TTC AT-3′). DNA sequencing by the Protein and Nucleic Acid Core Facility (Medical College of Wisconsin) confirmed the presence of the predicted sequences. Constructs lacking a His tag (sCD-MPR and E19Q/K137M) were cloned into the pPICZαA vector for expression in P. pastoris following methanol induction, whereas E19Q/K137MHis6 was placed into the pGAPZαA vector, which utilizes the glyceraldehyde-3-phosphate dehydrogenase promoter for constitutive expression of recombinant proteins in P. pastoris.
FIGURE 1. Amino acid sequence alignment of the extracytoplasmic region of the CD-MPR from various species.
The four N-glycosylation sites that have been mutated are indicated (*). The secondary structure of the sCD-MPR is shown above the sequence with a cylinder representing the α-helix and arrows represent β-strands. The cysteine residues are boxed in grey. Residues that are within hydrogen bonding distance to Man-6-P are shown: the four residues essential for Man-6-P binding are boxed in yellow, and residues which have been shown to have a modest (i.e., substitution of these residues results in <15-fold decrease in binding affinity (13)) effect on ligand binding are boxed in red. The residues involved in an inter-monomer salt bridge (Glu19 and Lys137) are boxed in green.
Expression and Purification of sCD-MPR, E19Q/K137M, and E19Q/K137MHis6 Constructs
cDNA constructs were linearized with either BspH1 (His-tagged constructs) or SacI (non-tagged constructs) and transformed into P. pastoris wild -type strain X-33 by electroporation. Zeocin-resistant transformants were selected on YPDS (1% yeast extract, 2% peptone, 2% dextrose, and 1 M sorbitol) following incubation at 30°C. Positive clones utilizing the pGAPαA expression vector were inoculated in liquid medium (1% yeast extract, 2% peptone, and 2% dextrose) and cultures were harvested after 3 days of growth at 30°C. Positive clones expressed using the pPICZαA system were first grown overnight at 28°C in minimal media (0.34% yeast nitrogen base without amino acids, 0.1 M potassium phosphate (pH 6.0), 38 mM ammonium sulfate, 4 × 10−5 % biotin, and 2% glycerol). To induce expression of the recombinant protein, the cells were resuspended to an OD600 of 1 in the above minimal media in which glycerol was replaced with 0.5% methanol as the sole carbon source and the cultures were harvested after 2–3 days of growth at 25°C. Following removal of the cells by centrifugation, medium containing E19Q/K137MHis6 was concentrated by filtration using Amicon stirred cells and then dialyzed extensively against metal binding buffer (20 mM Tris, 0.5 M NaCl and 10 mM imidazole, pH 8.0). After dialysis, the medium was passed over a nickel (Ni-NTA) affinity column. After washing, the column was eluted in a stepwise fashion using elution buffer (20 mM Tris, 0.5 M NaCl, pH 8.0) containing 20 mM, 50 mM, 100 mM or 250 mM imidazole. Purification of the non-tagged proteins (sCD-MPR and E19Q/K137M) involved dialyzing the concentrated medium against 50 mM imidazole (pH 6.5) and 150 mM NaCl prior to gel filtration using a Superdex 75 column (1.6 cm × 60 cm, GE Healthcare). The concentration of the purified protein was determined using the Bradford assay (BioRad) with bovine serum albumin (BSA) as the standard.
Isotopically labeled protein was generated by substituting ammonium sulfate with 15N-ammonium sulfate in the induction medium and used for NMR spectroscopic analyses. NMR spectral quality was dramatically improved by removal of the receptor’s single N-glycan at position 81: purified protein was treated with endo-β-N-acetylglucosaminidase H (endo H, 5mU, Roche Molecular Biochemicals) overnight and the deglycosylated protein was again subjected to gel filtration using a Superdex 75 column to remove the majority of the cleaved oligosaccharide. Residual oligosaccharide, along with contaminating mannan, which is secreted by P. pastoris yeast, was removed by Con A Sepharose chromatography. Proteins for NMR analyses were then buffer exchanged into a solution containing 10 mM deuterated bis-Tris (pH 6.5) and 150 mM NaCl.
Purification of Human β-Glucuronidase
Human β-glucuronidase was collected from serum-free conditioned medium from cells that over-express and secrete this lysosomal enzyme (MTX 3.2 cells were generously provided by Dr. William Sly, St. Louis University School of Medicine, St. Louis, MO). β-glucuronidase was purified by affinity chromatography on a CI-MPR Affigel-10 column to remove nonphosphorylated enzyme as described previously (16).
Biosensor Studies
All SPR measurements were performed at 25°C using a Biacore 3000 instrument (GE Healthcare). CM5 research grade sensor chips, surfactant P20 and amine coupling kits were also obtained from GE Healthcare. Purified proteins (sCD-MPRHis, E19Q/K137M) were immobilized on CM5 sensor chips following activation of the surface using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide as recommended by the manufacturer. Briefly, the proteins were injected onto the activated dextran surface at a concentration of 10–20 μg/ml in 10 mM sodium acetate buffer, pH 5.0 using immobilization buffer (10 mM MES pH 6.5, 150 mM NaCl and 0.005% (v/v) P20) as the running buffer. After coupling, unreacted N-hydroxysuccinimide ester groups were blocked with ethanolamine. The reference surface was treated in the same way except that protein was omitted. β-glucuronidase was prepared in running buffer (50 mM MES pH 6.5, 150 mM NaCl, 10 mM MnCl2, 5 mM β-glycerophosphate, and 0.005% (v/v) P20) and was injected in a volume of 80 μl over the coupled and reference flow cells at a flow rate of 40 μl/min. After 2 min, the solution containing β-glucuronidase was replaced with buffer and the complexes were allowed to dissociate for 2 min. The sensor chip surface was regenerated with a 25 μl injection of 10 mM Man-6-P at a flow rate of 10μl/min. The surface was allowed to re-equilibrate in running buffer for 1 min prior to subsequent injections. The sensorgrams were evaluated using the BIAevaluation software package (version 4.0.1) and the Kd determined from a 1:1 Langmuir binding model (kd/ka). All response data were double-referenced (17), where controls for the contribution of the change in refractive index were performed in parallel with flow cells derivatized in the absence of protein and subtracted from all binding sensorgrams.
Protein crystallization
The purified E19Q/K137MHis6 and E19Q/K137M mutants were dialyzed extensively against buffer containing 50 mM imidazole (pH 6.5), 150 mM NaCl, 10 mM MnCl2, and 5 mM β-glycerophosphate. E19Q/K137MHis6 was preincubated with 10 mM Man-6-P for 1 hr on ice prior to use in crystallization experiments. Crystallization was carried out at 19°C by vapor diffusion using the hanging drop method (18) by mixing equal volumes of the protein (13–15mg/ml) solution with the well solution (E19Q/K137MHis6: 0.1 M cacodylate pH 6.5, 25% polyethyleneglycol (PEG) 2000 monomethylether, 0.2 M ammonium acetate, 0.025 M octyl-β-D-glucopyranoside (only in the drop) or E19Q/K137MsulfatepH7.0: 0.1 M HEPES pH 7.0 and 3 M ammonium sulfate or E19Q/K137MsulfatepH6.5: 0.1 M cacodylate pH 6.5 and 3 M ammonium sulfate). The E19Q/K137MHis6 crystal was flash frozen after passing through a cryoprotection solution of the well solution with an additional 3% PEG as well as 20% glycerol added. The crystals from ammonium sulfate conditions were directly flash frozen without any additional cryoprotectant. All diffraction data were collected at 100°K using an R-AXIS IV++ equipped with a MicroMax007 generator and an Osmic mirror set. Data sets were processed with HKL2000 (19).
Structure determination
The structures were solved using Amore in CCP4 (20) (E19Q/K137MHis6) or Molrep (E19Q/K137MsulfatepH6.5) in CCP4i (21) using one monomer of the dimeric structure of the ligand-bound form of CD-MPR (Protein Data Bank code 1C39) as the search model. In the case of E19Q/K137MsulfatepH6.5, the starting search model did not include residues 133-141 (loop D). E19Q/K137MsulfatepH7.0 was solved by the difference Fourier method using the E19Q/K137M model with residues 133-141 (loop D) omitted. Resulting models were refined using CNS (22) in conjunction with manual model adjustments using Turbo-Frodo (23). In the case of E19K137MsulfatepH6.5, the twinning fraction (α = 0.284) and twinning operator (h, −k, −l) were calculated using CNS. The refinement was carried out again using CNS following protocols for treating partial hemihedral twinning. Data collection and refinement statistics for the final models are given in Table I.
Table 1.
Data collection and refinement statistics
| Glycosylated | endoH treated/conA purified | ||
|---|---|---|---|
| Crystal PDB ID |
EQKM/Man-6-P 3K41 |
EQKM/sulfatepH7.0 3K42 |
EQKM/sulfatepH6.5 3K43 |
| pH | 6.5 | 7.0 | 6.5 |
| Ligand/Mna | M6P | - | - |
| Data Collection | |||
| Space Group | P212121 | I4 | b I4 |
| Cell Dimensions: a, b, c (Å) | 53.2, 77.1, 79.9 | 102.6, 102.6, 100.1 | 103.0, 103.0, 99.8 |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Resolution (Å) | 50.0–1.90(1.97–1.90) | 50–2.3 (2.38–2.3) | 50–2.0 (2.07–2.0) |
| Data collection temp (°C) | −175 | −175 | −175 |
| Rsym | 0.13 (0.53) | 0.093 (0.481) | 0.068 (0.41) |
| I/σI | 26.7 (3.6) | 20.4 (3.7) | 24.7 (4.6) |
| Completeness (%) | 99.8 | 99.9 (100.0) | 99.7 (100.0) |
| Redundancy | 4.2 | 5.2 | 5.7 |
| Refinement | |||
| Resolution (Å) | 50–1.90 | 50–2.3 | 50–2.0 |
| No. of Reflections | 25567 | 22239 | 344841 |
| Rwork/Rfree | 21.6/25.5 | 22.8/24.8 | 16.5/19.7 |
| monomers/a.u. | 2 | 2 | 2 |
| Residues present in each monomer | A4-9, 16-154 B4-154 |
A3-10, 15-133, 141-154 B5-10, 15-154 |
A3-10, 15-133, 141-154 B5-10, 16-154 |
| Total No. residues/B-average (Å2) | |||
| Protein | 304/26.2 | 287/30.3 | 286/29.8 |
| M6P | 2/22.5 | - | - |
| c NAG(Man) | 4(2)/49.6 | 2/47.9 | 2/35.0 |
| water | 161/33.9 | 187/36.1 | 199/35.6 |
| sulfate | - | 8/59.2 | 3/54.1 |
| imidazole | 0 | 0 | 1/39.4 |
| acetate | 0 | 0 | 1/39.6 |
| Glycerol 1-phosphate | 0 | 2/47.2 | 0 |
| RMS deviations | |||
| Bond lengths (Å) | 1.29 | 1.2 | 1.2 |
| Bond angles (°) | 0.006 | 0.006 | 0.006 |
| Ramachandran plot (No. residues/%) | |||
| Most Favorable | 221/85 | 200/80.0 | 213/85.5 |
| Additionally Allowed | 39/15 | 48/19.2 | 36/14.5 |
| Generously Allowed | 0/0 | 1/0.4 | 0/0 |
| Disallowed | 0/0 | 1/0.4 | 0/0 |
Although Mn2+ was present in the crystallization media for 120805 and 041509 crystals, no appreciable Mn2+ peak was found in either structure.
Twinned with <I2>/<I>2 = 1.645, twinning fraction of 0.284, and twinning operator (h, −k, −l)
Oligosaccharide attached to Asn81; NAG, N-acetylglucosamine
Determination of Binding Affinities by Heteronuclear NMR Spectroscopy
NMR spectra were acquired on a Bruker 500MHz spectrometer equipped with a triple-resonance CryoProbe™ and processed with NMRPipe software (24). 15N sCD-MPR and 15N E19Q/K137M were titrated with incremental additions of Man-6-P and monitored via two-dimensional (2D) 15N-1H heteronuclear single quantum coherence (HSQC). Amide 1H-15N chemical shift perturbations were computed as where ΔδH and ΔδN are the changes in backbone amide 1H and 15N chemical shifts, respectively, in parts per million. Concentration-dependent changes in amide chemical shift perturbations were fitted to the following equation that accounts for ligand depletion:
where Δδ represents the chemical shift perturbation, Δδmax is the maximum chemical shift perturbation at 100% bound receptor, Kd is the apparent Man-6-P dissociation constant, and χ is the Man-6-P concentration. Kd and Δδmax values were obtained for each of the receptor residues by nonlinear curve fitting as previously described (25).
RESULTS AND DISCUSSION
Mutant Form of the sCD-MPR Lacking an Inter-Monomer Salt Bridge Exhibits Decreased Binding Affinity to Lysosomal Enzymes
Previous comparative structural analyses of the sCD-MPR identified a solvent accessible inter-monomer salt bridge (Glu19 from the N-terminal α-helix of one monomer and Lys137 from loop D of the other monomer) that was hypothesized to undergo protonation at acidic pH values, causing changes in quaternary structure and initiating a cascade of events which ultimately ends in the release of ligand (12). The observation that the region preceding Glu19 becomes disordered in structures solved under acidic pH conditions is consistent with this hypothesis (12). Furthermore, Glu19 and Lys137 are conserved in all species, with the exception of zebrafish in which Glu19 is replaced with Gln, and killfish and salmon in which Lys137 is conservatively replaced with Arg (Fig. 1). To determine whether the absence of this inter-monomer salt bridge affects the binding affinity of the receptor towards a lysosomal enzyme, β-glucuronidase, surface plasmon resonance (SPR) analyses were performed. The results demonstrate that sCD-MPR bound β-glucuronidase with high affinity (Kd = 26 nM) (Fig. 2A), whereas no detectable binding was observed to E19Q/K137M at concentrations as high as 120 nM (Fig. 2B). Consistent with these results, no detectable binding of E19Q/K137M was observed to a resin containing coupled phosphomannosyl residues (data not shown). In addition, two-dimensional 15N-1H HSQC NMR spectroscopy, which was used to measure the chemical shift perturbations upon the addition of Man-6-P, revealed that EQKM bound Man-6-P with an ~150-fold lower affinity than sCD-MPR (see below). Taken together, the results show that the mutant’s affinity for Man-6-P and β-glucuronidase is drastically compromised, demonstrating the essential role of the inter-monomer salt bridge in the ability of the CD-MPR to bind lysosomal enzymes with high affinity.
FIGURE 2. Interaction of sCD-MPR constructs with a lysosomal enzyme.
SPR analysis of β-glucuronidase binding to immobilized sCD-MPR (A) and E19Q/K137M (B) constructs. sCD-MPR and E19Q/K137M were immobilized on the surface of a CM5 sensor chip to a level of 800 and 770 response units, respectively. β-glucuronidase was injected in a volume of 80 μl over the sensor surface at a rate of 40 μl/min. After 2 min, the solution containing β-glucuronidase was replaced with buffer and the complexes were allowed to dissociate for 2 min. Various concentrations of β-glucuronidase (8, 10, 20, 40, 80, 100, and 120 nM) were injected over the immobilized receptors and the resulting sensorgrams are shown.
Crystallization of a Mutant Form of the sCD-MPR Lacking an Inter-Monomer Salt Bridge
Two versions (i.e., with and without a C-terminal His6 tag) of the mutant protein lacking an inter-monomer salt bridge (E19Q/K137MHis6 and E19Q/K137M) were crystallized and their structures determined (see Table I). The presence of the His tag had no significant affect on the structure of the receptor (see below). Consistent with this observation are previous studies in which we demonstrated that sCD-MPRHis6 bound the lysosomal enzyme, β-glucuronidase, with an affinity (Kd = 1.5 ± 0.2 nM) (13) which is comparable to that of the non-tagged version of the receptor (sCD-MPR) expressed in P. pastoris (Kd = 1.4 nM) (26) and of the full-length receptor isolated from mammalian tissues (Kd = 0.28 nM, (7); Kd = 4–5 nM, (27)). These results demonstrate that the presence of a C-terminal His tag does not alter the function of the sCD-MPR. Thus hereafter, we refer to E19Q/K137MHis6 and E19Q/K137M simply as “EQKM”.
Structures of two forms of EQKM have been obtained: 1) EQKM complexed with Man-6-P (EQKM/Man-6-P), and 2) EQKM crystallized in the absence of Man-6-P at pH 6.5 and pH 7.0. Each crystal form has one dimeric molecule in the asymmetric unit. As seen in other reported sCD-MPR structures (10–12, 28), the overall fold of each monomer relative to the other monomer in the asymmetric unit is the same, with the r.m.s. deviation ranging from 0.25 Å to 0.32 Å for 122 to 143 Cα atoms. However, as observed with some previously determined sCD-MPR structures (10–12), one (monomer B) of the two monomers has fewer disordered residues. Thus, unless otherwise stated, monomer B was used for subsequent analyses.
Comparison of the Monomer Structure of EQKM/Man-6-P and sCD-MPR
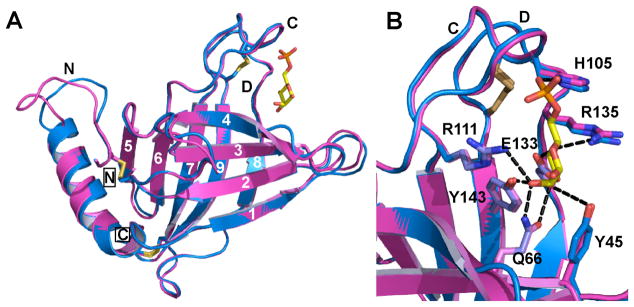
The overall topology of the monomer polypeptide of EQKM/Man-6-P consists of a flattened β-barrel comprised of nine β-strands, plus a short α-helix located at the amino terminus (Fig. 3A). Comparative analyses revealed that the structure of the EQKM/Man-6-P monomer is essentially the same as the wild-type sCD-MPR monomer in the presence of ligand (r.m.s. deviation of 0.32 Å for 122 Cα atoms) (Fig. 3A). Previous structures of the sCD-MPR have shown that the carbohydrate binding pocket contains six residues (Tyr45, Gln66, Arg111, Glu133, R135 and Tyr143) that are within hydrogen bonding distance to the mannose ring hydroxyl groups plus one residue (His105) that interacts with the phosphate moiety (Fig. 3B) (11, 28). These residues are conserved among species (Fig. 1) and mutagenesis studies have demonstrated that four of these residues (Gln66, Arg111, Glu133 and Tyr143) are essential for Man-6-P binding as substitution of any one of these residues results in ~1,000-fold decrease in the receptors’s binding affinity toward a lysosomal enzyme (13, 15). The architecture of the binding pocket, including the relative positions of these seven residues, is also preserved in the EQKM mutant in the presence of Man-6-P (Fig. 3B). The only appreciable difference between the two monomer structures is the conformation of the N-terminal loop (Leu8 -Lys14) and the first two amino acids (Glu15 and Ser16) of the α-helix that contains Gln19. However, the loop containing Lys137 (loop D, residues Glu133 - Cys141) maintains the same conformation as wild-type sCD-MPR (Fig. 3A). These comparative analyses indicate that disruption of the salt bridge between a residue in the N-terminal region and its partner located in loop D of the adjacent subunit does not influence the overall architecture of the Man-6-P binding pocket. Thus, the observed decrease in binding affinity towards lysosomal enzymes by EQKM (Fig. 2) does not appear to be due to a restructuring of the binding pocket, but must arise from how one monomer associates with the other to form the dimer.
FIGURE 3. Superimposition of the monomer structure of the EQKM mutant and sCD-MPR.
(A) The monomers of EQKM (cyan, 3K41) and sCD-MPR (magenta, 2RL8) crystallized in the presence of Man-6-P were compared. Loops N, C and D are labeled and the location of Man-6-P (yellow ball-and-stick) is shown. The N- and C-terminus are boxed. β-strands are numbered from 1 to 9. (B) Close-up view of the binding pocket. The seven amino acids involved in Man-6-P binding are labeled. Potential hydrogen bond interactions with Man-6-P are shown with black dashed lines.
Comparison of the Dimer Structure of EQKM/Man-6-P and sCD-MPR
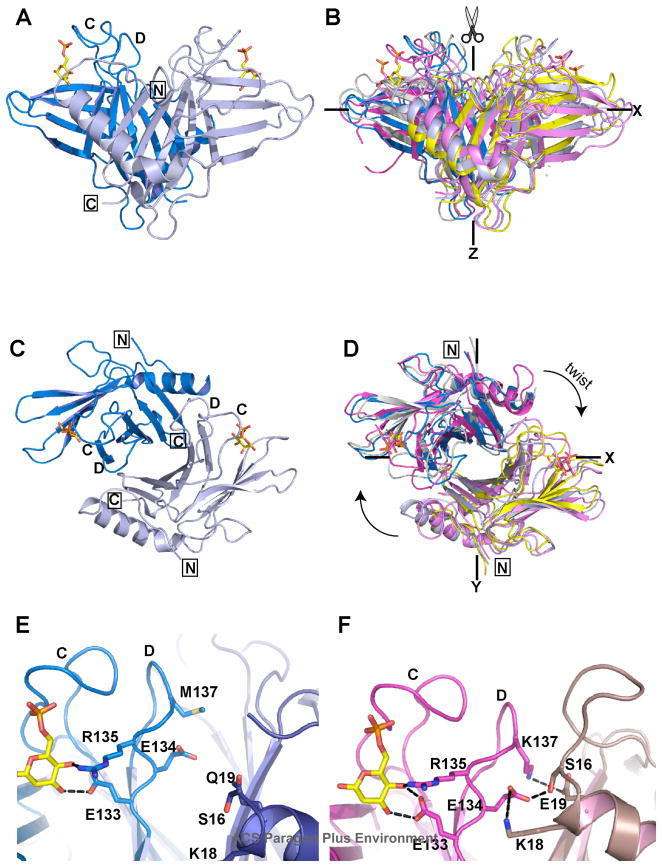
The CD-MPR functions as a dimer in the cell. Previous crystallographic studies have shown that the relative orientation of the monomers to each other is dependent upon the presence or absence of ligands, rather than pH (12). In the absence of ligand, the receptor adopts a ‘closed’ conformation with a reduced dimer interface area (1000 Å2 open versus 770 Å2 closed) that is due to loss of contacts between residues in loop D and residues near the N-terminal (residues Val9, Gly10, Ser16-Lys18) of the other monomer. Interestingly, the ligand-bound structure of the salt-bridge mutant (EQKM/Man-6-P) adopts a dimer conformation (Figs. 4A & 4C) that appears to be “in-between” the previously described closed (ligand-free) and open (ligand-bound) structures of the sCD-MPR (Figs. 4B & 4D). Specifically, the monomers of EQKM/Man-6-P are oriented with an ~20° scissor motion (10°/blade) away from the 2-fold axis (Z axis) on the XZ plane (Fig. 4B) and an ~9° clockwise twist motion relative to that of the ligand-unbound form (Fig. 4D). Relative to the ligand-bound form of the sCD-MPR, the monomers of EQKM/Man-6-P are oriented with an ~16° scissor motion (8°/blade) toward the 2-fold axis (Z axis) on the XZ plane (Fig. 4B) and an ~9° counter-clockwise twist motion (Fig. 4D). Although the ligand binding pocket architecture in the EQKM/Man-6-P structure is identical to that of sCD-MPR (Fig. 3B), the interactions between the residues of the binding pocket and the neighboring residues that support the binding pocket are very different. The sCD-MPR structure reveals that the binding cavity is stabilized by interactions between loop D of one monomer and the residues within the N-terminal region of the other monomer, which includes electrostatic interactions/hydrogen bonds (Glu19-Lys137, Lys18-Glu134, and Ser16-Glu134) (Fig. 4F). The absence of the salt bridge, Glu19–Lys137, in EQKM disrupts these inter-monomer ionic interactions resulting in a less stable binding pocket (Fig. 4E), which in turn results in a >100-fold decrease in EQKM’s ligand binding affinity (see Figs. 2 and 5B).
FIGURE 4. Comparison of the dimer structures of sCD-MPR in the absence and presence of Man-6-P with that of EQKM/Man-6-P.
(A) The Man-6-P bound structure of EQKM (3K41, blue/grey) was overlayed (B) with the structures of sCD-MPR crystallized in the unbound state (2RL7, grey/yellow) as well as bound to Man-6-P (2RL8, magenta/pink). (C) and (D), The dimers have been rotated down 90° from the view in (A) and (B), showing the superimposition looking down the Z axis. Close-up view showing the dimer interface of (E) EQKM/Man-6-P (3K41) and (F) sCD-MPR (2RL8). Residues on loop D proposed to interact with either the ligand or loop N of the other monomer are shown. Black dashed lines indicate potential hydrogen bond or salt bridge interactions. Loops C and D are indicated in panels A, C, E and F. The N- and C-terminus are boxed in panels A, C and D. The location of Man-6-P (yellow ball-and-stick) is shown.
FIGURE 5. Effect of elimination of the inter-monomer salt bridge (Glu19 – Lys137) on the solution structure and Man-6-P binding affinity of sCD-MPR.
(A) Overlay of 1H-15N HSQC spectra collected on sCD-MPR in the presence (grey) and absence (magenta) of Man-6-P. The 1H-15N HSQC spectrum of EQKM (cyan) collected in the absence of Man-6-P is shown for comparison. The side chain amide group (Gln19) of EQKM is indicated by the dashed grey circle. (B) Apparent binding affinities (Kd) were obtained by 1H-15N HSQC spectroscopy titrating Man-6-P with 15N-labeled sCD-MPR or EQKM and monitoring chemical shift perturbations for three different nitrogen groups.
Influence of Ligand Binding on the Solution Structure of EQKM and sCD-MPR
To further investigate the dynamic nature of the CD-MPR as inferred by the various crystal structures of the receptor (Figs. 4B & 4D), the solution structure of sCD-MPR and EQKM were investigated by NMR spectroscopy. Two-dimensional 15N-1H HSQC spectra were collected for sCD-MPR and EQKM in the presence and absence of Man-6-P and compared. Inspection of the sCD-MPR spectrum indicates that the receptor is dynamic in the absence of Man-6-P as evidenced by the heterogeneous peak intensities and line broadening observed for most of the resonances (Fig. 5A). In contrast, binding of Man-6-P restricts the sCD-MPR to the “open” conformation as shown by the homogeneous peak intensities and line widths (Fig. 5A). Additionally, a comparison of the sCD-MPR and EQKM spectra collected in the absence of Man-6-P revealed that the peaks in the spectrum overlay well, suggesting that EQKM can adopt the “closed” conformation similar to the sCD-MPR (Fig. 5A). Titrations of 15N labeled sCD-MPR or 15N labeled EQKM with Man-6-P were monitored by 15N-1H HSQC spectroscopy to identify amino acid residues that experienced chemical shift changes upon Man-6-P binding. Although chemical shift assignments have not yet been made, relative chemical shift changes were monitored for three different backbone amides for sCD-MPR, and two different backbone amides as well as the side chain amide of Gln19 for EQKM. Nonlinear fitting of the chemical shift changes as a function of Man-6-P concentration yielded apparent Kd values of ~1 mM and ~150 mM for sCD-MPR and EQKM, respectively (Fig. 5B). These results demonstrate that EQKM binds Man-6-P with an ~150-fold lower affinity than the wild-type receptor, which is consistent with the data obtained by SPR analyses (Fig. 2).
Crystal Structure of EQKM in the Absence of Man-6-P
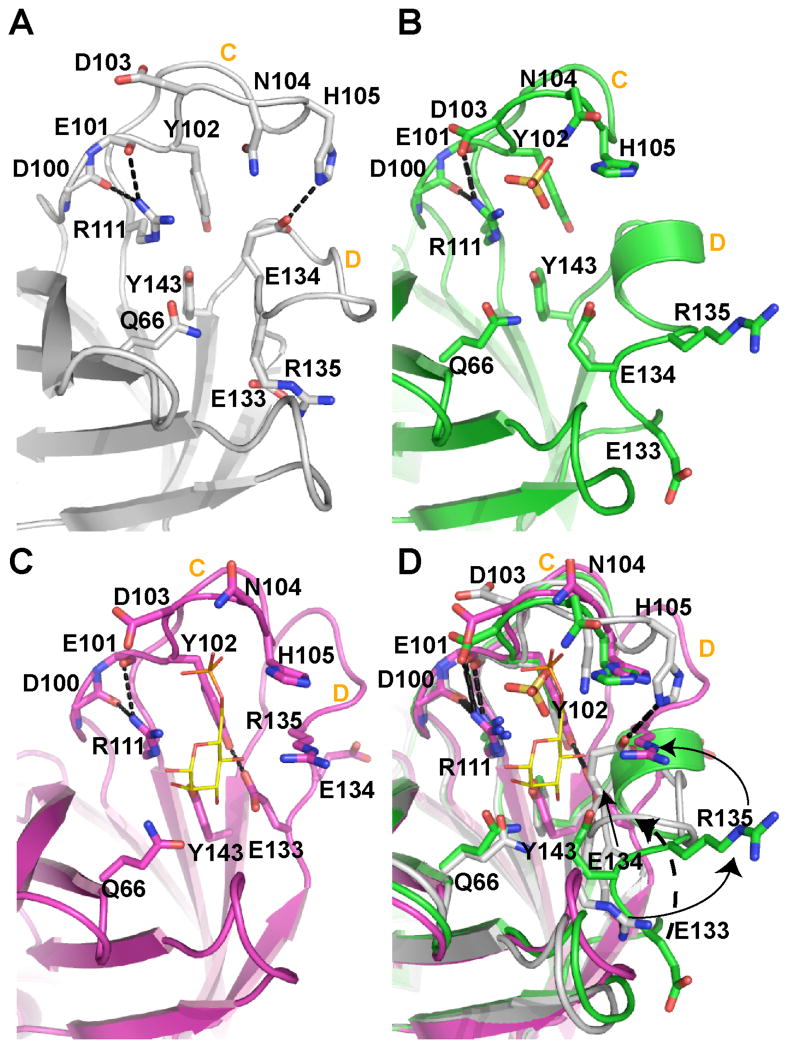
Given the observed differences in the quaternary structure of EQKM bound to Man-6-P (Figs. 4B & 4D), we sought to obtain the structure of EQKM in a ligand-free form by crystallizing the protein in the absence of Man-6-P at pH 6.5. The resulting structure was found to contain a sulfate ion in the ligand binding cavity near where the phosphate moiety of Man-6-P would reside (Fig. 6B). The presence of sulfate was due to the crystallization conditions in which 3 M ammonium sulfate was used as the precipitant. The protein was crystallized at a higher pH (pH 7.0) in an attempt to deprotonate His105 (which coordinates the phosphate group) in order to eliminate binding of the sulfate ion. However, both resulting structures were essentially identical with an r.m.s. deviation of 0.17 Å and in both structures the residues within loop D were disordered in one of the two monomers. Therefore, detailed structural analysis was carried out only on the structure crystallized at pH 7.0 (EQKM/sulfate). Interestingly, although the relative orientation of the monomers of this sulfate bound structure is the same as that found in EQKM/Man-6-P, the well defined monomer demonstrates some intriguing differences that have not been seen in any of the sCD-MPR structures solved to date. The major difference resides in the conformation of loop D. Our previous structures of sCD-MPR (10, 12) demonstrated that in the ligand-free state loop D pivots into the carbohydrate binding site and establishes a network of hydrogen bond interactions which preserve the orientation of three (Gln66, Arg111, Tyr143) out of the four essential binding site residues: only the position of Glu133 is altered since it resides within loop D (Fig. 6A). In contrast, in the presence of ligand, the N- and C-terminal residues of loop D form a side of the binding pocket near the mannose ring while the residues at the tip of the loop (encompassing Lys137) interact with residues in the N-terminal region of the adjacent monomer (including Glu19) (Figs. 4F & 6C) (10, 12). Importantly, these interactions form a direct link between ligand binding in one monomer and the N-terminal region of the other monomer (Fig. 4F). The EQKM/sulfate structure reveals that the residues of loop D form one turn of an α-helix (Fig. 6B). Although the significance of this short helical conformation is not entirely clear, the observation that loop D adopts a different conformation (either disordered or α-helical) (Fig. 6B) suggests that this region of the receptor is highly mobile and its reordering plays a role in the dimer reorientation observed in response to the presence or absence of ligand.
FIGURE 6. Close-up view of the binding site unoccupied, partially occupied, or fully bound to ligand.
(A) In the complete absence of ligand, loop C is elongated to form a β-finger (sCD-MPR, 2RL7). (B) However, loop C contracts in the presence of a sulfate ion (yellow ball-and-stick) (EQKM/sulfate, 3K42). The presence of sulfate also results in the movement of loop D out of the binding pocket and the reordering of loop D to form an intermediate α-helix. (C) The occupation of both the phosphate and mannose binding regions of the receptor (sCD-MPR, 2RL8) transitions the residues of loop D to the side of the binding pocket. (D) Superimposition of all three states of the binding pocket. Residues are labeled as well as loops C and D in all four panels.
Presence of a Sulfate Ion in the Binding Site of EQKM/sulfate Mimics an Intermediate Binding Site Conformation
The structure of the sCD-MPR under a variety of conditions (10–12, 28) has provided insight into the mechanism of ligand binding and release. The binding pocket of CD-MPR is comprised of two functional regions: the phosphate binding area (composed of residues in loop C) and the mannose binding area (composed of residues from β-strands 3 and 4 of the front sheet as well as residues near the N- and C-terminus of loop D. As described above, in the absence of ligand, loop D is positioned in the carbohydrate recognition site, and the residues of loop C also undergo movement relative to each other (Fig. 6A) allowing for changes in the interplay between the two functional regions of the binding pocket. In the ligand bound structures of the sCD-MPR (Fig. 6C), Tyr102 is within hydrogen bonding distance to Glu133; however, the interaction of Glu133 shifts from Tyr102 to Arg135 in the ligand-free state (Fig. 6A), and Glu134 of loop D is now in position to hydrogen bond to His105 of loop C (Fig. 6A). The shifting of the hydrogen bonding pattern within the binding site (Figs. 6A, 6C & 6D) allows the side chain of Asp103 (coordinates the cation Mn+2) to move out of the phosphate binding area, thus opening the cavity to allow subsequent access of a phosphate moiety. Taken together, these interactions serve to link the two regions of the binding pocket together.
The EQKM/sulfate structure captures a unique view of the relationship between the phosphate and mannose binding regions by allowing only half of the binding site to be occupied (i.e., the phosphate recognition region) (Fig. 6B). In this location, it is possible for the sulfate ion to have electrostatic interactions with the side chain nitrogen atoms of His105, Asn104 and Lys73 (Fig. 6B). Therefore, in terms of loop C, the EQKM/sulfate structure more closely resembles a bound structure. As previously reported for other bound and unbound structures, three of the four essential binding residues do not alter their position in the mannose binding area in the presence of sulfate. The residues in loop D (including Glu133), however, do not overlay with their counterparts in either the ligand-bound or ligand-free structures (Fig. 6D); rather, their position suggests a possible intermediate state between the ligand-bound and ligand-free states.
Monomer Reorientation and the Dimer Interface in the Presence or Absence of Ligand
As described above, the structure of sCD-MPR is dynamic and undergoes a series of transitions as ligand is bound and released, with the presence of ligand necessitating the movement of loop D residues out of the region occupied by the mannose ring (Fig. 6). In addition, changes in quaternary structure occur that can be described as a scissoring and twisting motion of one monomer relative to the other (Fig. 4B & 4D), resulting in changes in association between residues of the dimer interface. We have captured four states along this pathway of ligand binding: ligand-free, closed dimer conformation (state 1, wild-type sCD-MPR, 2RL7); partial occupancy, only the phosphate portion of the binding pocket is filled and has an intermediate dimer conformation (state 2, EQKM/sulfate, 3K42); both phosphate and carbohydrate regions of the binding site are occupied and has an intermediate dimer conformation (state 3, EQKM/Man-6-P, 3K41); and ligand-bound, open dimer conformation (state 4, wild-type sCD-MPR, 2RL8). The dimer interface is comprised of two regions: the interactions between loop N and loop D which are present only when ligand is present and 2) a set of core residues involved in monomer-monomer interactions that are present in both the ligand-free and ligand-bound conformations. This core region consists of hydrophobic and hydrophilic interactions between residues located in the C-terminal β-sheets as well as ionic interactions between residues near loop C of one monomer and loop D of the other. A comparison of the ligand unbound structure (state 1) and the partially occupied binding pocket (state 2) reveals that these interface residues adopt essentially the same orientation, with the exception of Ser132 (located on the C-terminus of strand 8 leading into loop D) and Asp140 (within loop D) (Fig. 7A). The formation of the helical turn in loop D results in a 1.9 Å repositioning of the Cα atoms of Asp140 while the Cα atoms of Ser132 differ by ~4 Å. The presence of the sulfate ion forces the residues of loop C into a bound conformation while the residues in loop D are left in an intermediate or transitional position. Addition of mannose to the binding pocket (Fig. 7B, state 3) continues the movement of Asp140 plus rotation of Phe142 is now observed. Comparison of both of these sets of structures reveals that backbone and side chain movements are fairly well confined to the area of the interface nearest loop D (Fig. 7A & 7B circled region). The relocation of the residues in loop D to form the binding pocket places Asp140 in a region of concentrated positively charged residues including Lys97 and Arg112 (Fig. 7G). The EQKM/Man-6-P structure reveals that Asp140 of one monomer is within ~6 Å of its counterpart in the other monomer (Fig. 7G). This juxtapositioning of negative charge may serve to destabilize this dimer conformation in this mutant form of the receptor (EQKM), thus contributing to its much lower (~150-fold) affinity for ligand. The final step to form the fully ligand-bound wild-type structure, with both regions of the binding site fully occupied (Fig. 7C, state 4), produces additional side chain rearrangements. However, these rearrangements are now localized to the N-terminal side of the interface and involve Gln84, Phe86 and Trp91. Comparison of these different structures illustrates the dynamic nature of the CD-MPR.
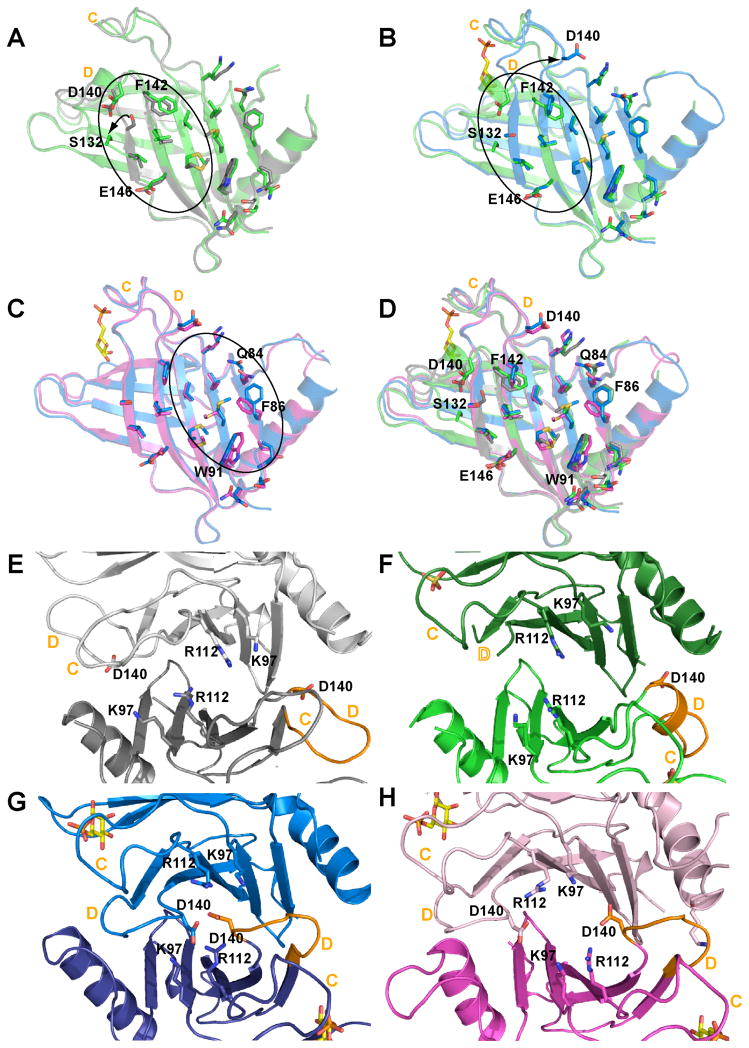
FIGURE 7. Mapping the changes in the dimer association for the four states of sCD-MPR.
(A) Overlay of unbound (2RL7, grey) and partially bound (3K42, green) receptor. The occupation of the phosphate binding region by sulfate (green) is accompanied by reordering of residues of loop D which displaces Ser132 out of β-strand 8 while causing more minor perturbations in the locations of Asp140 (unbound sCD-MPR, grey, 2RL7). (B) Overlay of the partially bound (3K42, green) and fully bound (2RL8, blue) mutant receptor. The presence of Man-6-P (EQKM/Man-6-P, 3K41, blue) results in the complete transition of loop D to that found in the fully bound structure. (C) Overlay of the Man-6-P bound mutant (3K41, blue) and wild-type (2RL8, pink) receptors. The final dimer transition following binding of ligand (2RL8, bound) involves shifting of the side chains nearer the N-terminal loop. (D) Overlay of all monomers. In panels A, B and C residues proposed to be involved the transition from the unbound to bound states are circled. For clarity the dimers were rotated downward 90° from panels A-D to show the electrostatic interactions near loops C and D of dimer in the (E) unbound (2RL7), (F) partially bound (i.e., phosphate portion of the binding site occupied with sulfate) (EQKM/sulfate, 3K42), (G) Man-6-P bound in absence of the Glu19-Lys137 salt bridge (EQKM/Man-6-P, 3K41) and (H) sCD-MPR (2RL8). Loops C and D are labeled in orange in all panels.
Conclusions and Future Studies
The CD-MPR travels between various cellular compartments and must undergo conformational changes between ligand-bound and unbound states during the loading and unloading of its cargo. We now have structures of sCD-MPR in four different conformations representing different stages of ligand binding: the wild type ligand-free structure (12) (PDB code, 2RL7), EQKM/sulfate (current work, PDB code 3K42), EQKM/Man-6-P (current work, PDB code 3K41), and the wild type fully bound structure (12) (PDB code, 2RL8). Thus, we are now in a position to better understand the individual steps involved in the inter-conversion of the ligand-free and ligand-bound states of the CD-MPR. When the CD-MPR is in the ligand-free state (state 1), the receptor adopts the “closed” form where loop D folds into the mannose binding site to interact with the residues that bind the sugar ring, thereby preserving the conformations of the sugar binding residues. The inter-monomer interactions between loops N (E19) and D (K137) are not present in this conformation. State 2 of the receptor is represented by the occupation of the phosphate binding region by the analogous tetrahedral sulfate anion. Although this state is artificial since the natural ligand would also have a carbohydrate moiety attached to the phosphate, it allows us to decouple the binding of carbohydrate from that of the phosphate group, thereby providing information about an intermediate state of binding by the receptor. When the phosphate moiety of the ligand binds to the receptor, loop C including Asp103, Asn104, and His 105 reconfigures and accommodates the phosphate of the ligand as seen in the structure of EQKM/sulfate. When loop C reorders, the interactions between it and the rest of the binding pocket are altered. These alterations may facilitate the movement of loop D out of the binding pocket through a series of reordering steps (one being the α-helical conformation captured in the EQKM/sulfate structures). The partial occupation of the binding site is accompanied by changes in the dimer to the intermediate state. In the third state of the receptor (EQKM/Man-6-P), both the phosphate and the carbohydrate recognition regions of the binding site are occupied and the four residues essential to carbohydrate recognition are in place. However in this state the receptor has not achieved optimal dimer orientation for maximal binding affinity. Further reorientation of the monomers occurs in the fourth state (representing the highest affinity state) to optimize inter-monomer interactions. As demonstrated by the characterization of the EQKM mutant presented in this work, these inter-monomer interactions including the salt bridge between Glu19 of one monomer and K137 of the other prove to be critical for the receptor to attain high affinity ligand binding.
It has been shown that the loading and unloading of its cargo are facilitated by changes of pH in each subcellular compartment where the CD-MPR travels (29). However, it is not clear which region or residue(s) of the CD-MPR is responsible for sensing these pH changes. We have hypothesized Glu133 (one of the ligand binding residues in loop D) and the phosphate moiety of Man-6-P are responsible for responding to the pH changes for unloading the ligand (12). However, there are other candidates that might act as the pH sensor(s), including His105 in loop C (binds to phosphate), Glu134 loop D, Glu19 near the N-terminus and Glu146 near the hydrophobic interface. Future studies using NMR and EPR spectroscopic analyses will be required to fully evaluate the dynamic nature of the CD-MPR, to assess its allosteric behavior, and to identify the residues involved in pH-dependent ligand binding.
Acknowledgments
The BIAcore 3000 instrument (Protein and Nucleic Acid Core Facility, Medical College of Wisconsin) was purchased through a grant from the Advancing a Healthier Wisconsin program.
The abbreviations used are
- CD-MPR
cation-dependent mannose 6-phosphate receptor
- CI-MPR
cation-independent mannose 6-phosphate receptor
- TGN
trans Golgi network
- Man-6-P
mannose 6-phosphate
- Ni-NTA
nickel nitriloacetic acid
- SPR
surface plasmon resonance
- sCD-MPR
soluble form of the CD-MPR containing a single N-glycosylation site at Asn81
- Glc-6-P
glucose 6-phosphate
- endo H
endo-β-N-acetylglucosaminidase H
- Con A
concanavalin A
- r.m.s
root-mean-square
- HSQC
heteronuclear single quantum coherence
Footnotes
This work was supported by National Institutes of Health Grant R01DK42667 (to N.M.D. and J.-J.P.K.)
References
- 1.Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–213. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 3.Dahms NM, Olson LJ, Kim JJ. Strategies for carbohydrate recognition by the mannose 6-phosphate receptors. Glycobiology. 2008;18:664–678. doi: 10.1093/glycob/cwn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein M, Zijderhand-Bleekemolen JE, Geuze H, Hasilik A, von Figura K. Mr 46,000 mannose 6-phosphate specific receptor: its role in targeting of lysosomal enzymes. EMBO J. 1987;6:2677–2681. doi: 10.1002/j.1460-2075.1987.tb02559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe H, Grubb JH, Sly WS. The overexpressed human 46-kDa mannose 6-phosphate receptor mediates endocytosis and sorting of beta-glucuronidase. Proc Natl Acad Sci USA. 1990;87:8036–8040. doi: 10.1073/pnas.87.20.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong PY, Kornfeld S. Ligand interactions of the cation-dependent mannose 6-phosphate receptor. Comparison with the cation-independent mannose 6-phosphate receptor. J Biol Chem. 1989;264:7970–7975. [PubMed] [Google Scholar]
- 9.Qian M, Sleat DE, Zheng H, Moore D, Lobel P. Proteomics analysis of serum from mutant mice reveals lysosomal proteins selectively transported by each of the two mannose 6-phosphate receptors. Mol Cell Proteomics. 2008;7:58–70. doi: 10.1074/mcp.M700217-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Olson LJ, Zhang J, Dahms NM, Kim J-JP. Twists and turns of the CD-MPR: ligand-bound versus ligand-free receptor. J Biol Chem. 2002;277:10156–10161. doi: 10.1074/jbc.M112230200. [DOI] [PubMed] [Google Scholar]
- 11.Roberts DL, Weix DJ, Dahms NM, Kim J-JP. Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor. Cell. 1998;93:639–648. doi: 10.1016/s0092-8674(00)81192-7. [DOI] [PubMed] [Google Scholar]
- 12.Olson LJ, Hindsgaul O, Dahms NM, Kim JJ. Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor. J Biol Chem. 2008;283:10124–10134. doi: 10.1074/jbc.M708994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun G, Zhao H, Kalyanaraman B, Dahms NM. Identification of residues essential for carbohydrate recognition and cation dependence of the 46-kDa mannose 6-phosphate receptor. Glycobiology. 2005;15:1136–1149. doi: 10.1093/glycob/cwi098. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Dahms NM. Site-directed removal of N-glycosylation sites in the bovine cation- dependent mannose 6-phosphate receptor: effects on ligand binding, intracellular targetting and association with binding immunoglobulin protein. Biochem J. 1993;295:841–848. doi: 10.1042/bj2950841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson LJ, Hancock MK, Dix D, Kim JJP, Dahms NM. Mutational analysis of the binding site residues of the bovine cation-dependent mannose 6-phosphate receptor. J Biol Chem. 1999;274:36905–36911. doi: 10.1074/jbc.274.52.36905. [DOI] [PubMed] [Google Scholar]
- 16.Marron-Terada PG, Brzycki-Wessell MA, Dahms NM. The two mannose 6-phosphate binding sites of the insulin-like growth factor-II/mannose 6-phosphate receptor display different ligand binding properties. J Biol Chem. 1998;273:22358–22366. doi: 10.1074/jbc.273.35.22358. [DOI] [PubMed] [Google Scholar]
- 17.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 18.McPherson A. Current approaches to macromolecular crystallization. Eur J Biochem. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative CPN. The CCP4 suite: programs for protein crystallography. Acta Crystallography Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallography. 2004;D60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallography. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Roussel A, Cambillau C. Biographics. 4.2. LCCMB; Marseille, France: 1994. TURBO-FRODO. The Manual. [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Vax A. NMRPipe: a multidemensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Seibert C, Veldkamp CT, Peterson FC, Chait BT, Volkman BF, Sakmar TP. Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases. Biochemistry. 2008;47:11251–11262. doi: 10.1021/bi800965m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy ST, Dahms NM. High-level expression and characterization of a secreted recombinant cation-dependent mannose 6-phosphate receptor in Pichia pastoris. Protein Expr Purif. 2002;26:290–300. doi: 10.1016/s1046-5928(02)00542-9. [DOI] [PubMed] [Google Scholar]
- 27.Ma ZM, Grubb JH, Sly WS. Cloning, sequencing, and functional characterization of the murine 46- kDa mannose 6-phosphate receptor. J Biol Chem. 1991;266:10589–10595. [PubMed] [Google Scholar]
- 28.Olson LJ, Zhang J, Lee YC, Dahms NM, Kim JJP. Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor. J Biol Chem. 1999;274:29889–29896. doi: 10.1074/jbc.274.42.29889. [DOI] [PubMed] [Google Scholar]
- 29.Imort M, Zuhlsdorf M, Feige U, Hasilik A, von Figura K. Biosynthesis and transport of lysosomal enzymes in human monocytes and macrophages. Effects of ammonium chloride, zymosan and tunicamycin. Biochem J. 1983;214:671–678. doi: 10.1042/bj2140671. [DOI] [PMC free article] [PubMed] [Google Scholar]