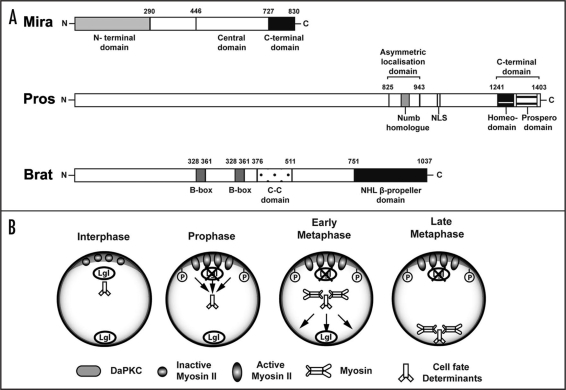
Figure 5.
Basal cell fate determinants and potential mechanism of asymmetric localization. (A) Cell fate determinant proteins and their functional domains. Miranda (Mira) protein consists of the central domain that provides binding sites for the cell fate determinants carrying them as its cargo proteins. The N-terminal domain of Mira is required for its association to the membrane, whereas, the C-terminal domain plays a role in cortical localization, and release of its cargo proteins upon cytokinesis. The Prospero (Pros) protein contains an asymmetric localization domain required for binding to Miranda. Within this region lies the Numb homolog domain, that may indicate a common asymmetric segregation mechanism shared with Numb. Other well established domains include the nuclear localizing signal (NLS), the homeodomain required for DNA binding, as well as the prospero domain that masks nuclear exclusion. Brain tumor (Brat) protein is characterised by the presence of two N-terminal Zinc binding B-boxes, a coiled-coil region, and the C-terminal NHL β-propeller domain, which can bind the central domain of Miranda. (B) Basal protein targeting of cell fate determinants. When Lgl is phosphorylated and inactivated by DaPKC at the apical cortex, Myosin II becomes activated around early prophase to exclude Mira and its cargo proteins from the cortex into the cytoplasm. Subsequently, Mira may bind to Myosin VI to be transported basally to form a crescent around metaphase. See text for details.