Abstract
Mu opioid receptor antagonists have clinical utility and are important research tools. To develop non-peptide and highly selective mu opioid receptor antagonist, a series of 14-O-heterocyclic-substituted naltrexone derivatives were designed, synthesized, and evaluated. These compounds showed subnanomolar-to-nanomolar binding affinity for the mu opioid receptor. Among them, compound 1 exhibited the highest selectivity for the mu opioid receptor over the delta and kappa receptors. These results implicated an alternative ‘address’ domain in the extracellular loops of the mu opioid receptor.
Keywords: Opioid, Mu opioid receptor, Antagonist, Naltrexone
Opioid receptors were generally classified into three subtypes based on the pharmacological, behavioral, and biochemical studies.1–3 Opioid antagonists have played very important roles in the study of opioid receptors. In fact, an agonist is characterized as opioid-receptor-mediated only if its effect is competitively inhibited by an opioid antagonist.4,5 It is important to have receptor-selective opioid antagonists as tools to identify the receptor types related to the interaction with opioid agonists.4–6 The mu opioid receptor (MOR) is the major type that mediates opioid analgesic effects of morphine, although all three opioid receptors can be involved in analgesia. The characterization of the MOR structure–function relationship is essential because it has been found that morphine’s analgesic effect, addictive properties, and other major side effects are abolished in MOR knock-out mice.7,8 Moreover, it has been demonstrated that the analgesic effects and the adverse side effects (including addiction and abuse liability) of morphine are primarily due to its interaction with the MOR.4 In fact, naltrexone, an opioid antagonist with moderate selectivity for the MOR, has been shown to block relapse and curb drug craving in post-dependent opiate addicts.9,10 Recent research results also indicate that MOR antagonists can be used in the treatment of obesity, psychosis and Parkinson’s disease.11 Furthermore, highly selective MOR antagonists can be used as probes to characterize the MOR-binding pocket. Yet the lack of a non-peptidyl, highly selective, and potent MOR antagonist limits our understanding of the structure–function relationship of the MOR, the interaction of non-peptidyl MOR agonists with the receptor, and more specifically, the activation mechanism of the receptor related to its role in drug abuse and addiction.
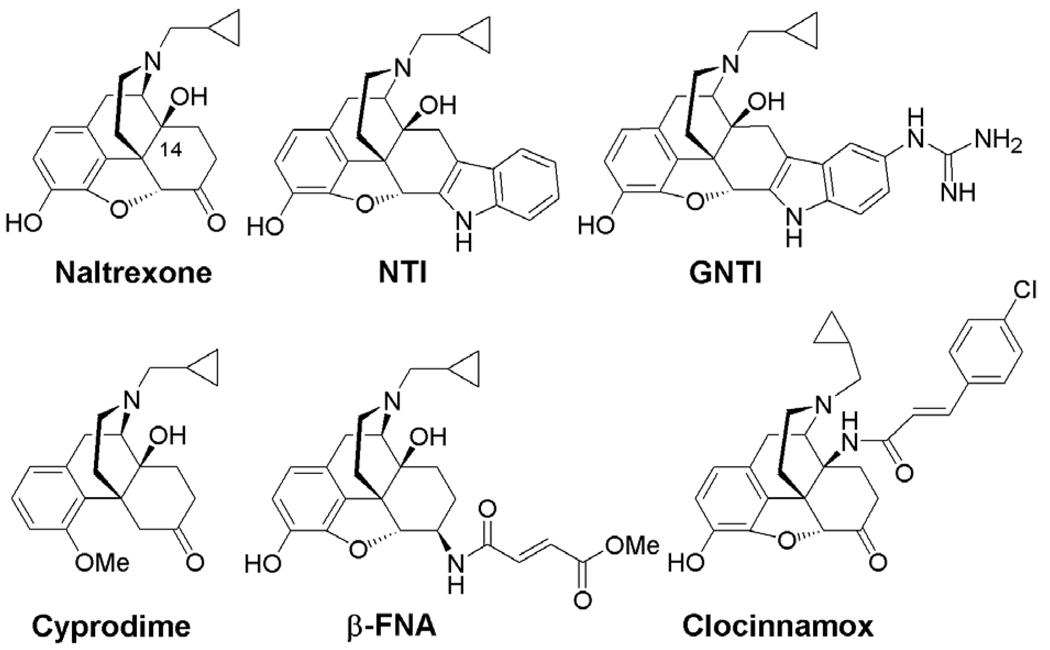
Schwyzer et al. proposed the ‘message-address’ concept in his analysis of the structure–activity relationship of ACTH, adrenocorticotropic hormone, and related hormones.12 By applying the ‘message-address’ concept, highly selective non-peptide antagonists for the kappa opioid receptor (KOR) (e.g., norbinaltorphimine (norBNI) and 5′-guanidinonaltrindole (GNTI)),13,14 and for the delta opioid receptor (DOR) (e.g., naltrindole (NTI))15 were designed and synthesized several years ago (Fig. 1). Thus far no potent and highly selective antagonist derived from morphinan’s structural skeleton has been developed for the MOR, although some moderately potent ligands, for example, cyprodime,16 are available. Compared with the high selectivity of GNTI for the KOR (Ki value ratios are mu/kappa ≈ 120, delta/kappa ≈ 250)14 and NTI for the DOR (Ki value ratios are mu/delta ≈ 152, kappa/delta ≈ 276),15 cyprodime only has a moderate selectivity for the MOR over the DOR and KOR (Ki value ratios are kappa/mu ≈ 45, delta/mu ≈ 40).16a At the same time, β-funaltrexamine (β-FNA), clocinnamox, and other compounds, act as selective but irreversible antagonists for the MOR.17 Therefore the development of a highly selective, non-peptidyl, and reversible MOR antagonist is highly desired.
Figure 1.
Morphinan derivatives as opioid selective antagonists.
It was reported that the extracellular loop (EL) domains of the MOR are critical for the binding of MOR-selective agonists, such as morphine, sufentanil, lofentanil, and DAMGO.18 At the same time, site-directed mutagenesis studies have revealed that certain amino acid residues in this domain may be essential for ligand (including agonist and antagonist) selectivity for the MOR over the other two opioid receptor types.19 Therefore, a non-peptide ligand with potential interaction with the EL domains of the MOR, would be favorable for its selectivity for the MOR.
Due to the lack of the crystal structure of the MOR, so far most molecular design efforts directed toward development of selective opioid ligands have been based on structure–activity relationship studies. As a matter of fact, in the entire superfamily of GPCRs, only the X-ray crystal structures of bovine rhodopsin,20–23 opsin,24 and the human β2-25–28 and β1-adrenergic receptors29 have been successfully obtained with high resolution. Thus far, most of the molecular models of other GPCRs have been constructed using rhodopsin’s structure as a template via homology modeling. Homology modeling of GPCRs has been successfully applied to further understand ligand–protein interactions, and to identify new and potent ligands. It is believed that with all the lessons learned from previous experience, GPCR homology modeling based on the bovine rhodopsin X-ray crystal structure can aid in structure-based drug design and virtual screening for therapeutic applications.30–37 For example, a homology model of the Angiotensin II Type 1 (AT1) receptor was used to further explore the binding sites of several non-peptide AT1 receptor antagonists.38 A homology model of the M1 muscarinic acetylcholine receptor was applied to understand the mechanism by which the agonist–receptor complex activates G proteins.39
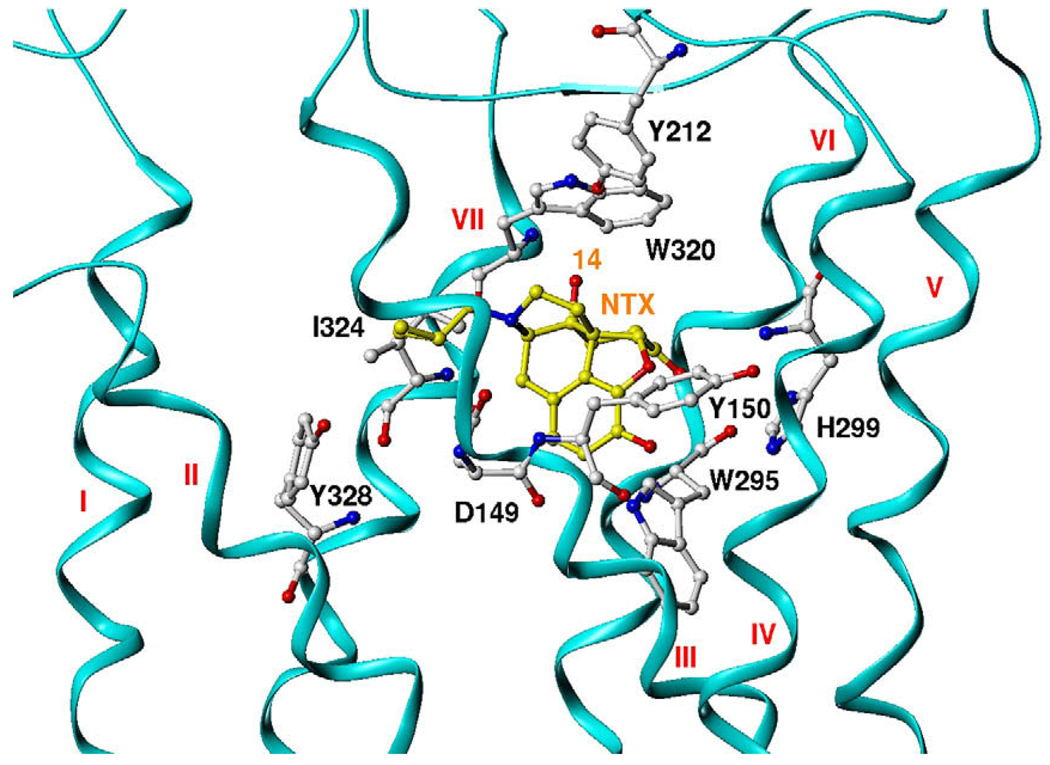
Recently, we reported the construction of a MOR homology model based on the crystal structure of bovine rhodopsin.40 This model contained not only the transmembrane helical domains, but also the extracellular and intracellular loops so that the model we obtained was integrated and complete. This model was further optimized in a membrane-aqueous system by molecular dynamics simulations. Similar homology models of the DOR and KOR were then constructed (see Supplementary information for details). Naltrexone is an ideal template for the design of selective MOR antagonists, because it has subnanomolar-to-nanomolar affinity for all three opioid receptor types and shows moderate selectivity for the MOR over the other two opioid receptor types. Figure 2 shows that in a representative binding mode of naltrexone in the MOR, the 14-hydroxyl group of naltrexone is pointing to the EL3 loop and the upper-level region of TM6/7. Compared to the amino acid residues in the corresponding domains of the KOR and DOR, some non-conserved residues, for example, Tyr212 and Trp320, in MOR could act as hydrogen bonding donor/acceptors. This unique feature in the MOR antagonist-binding locus might form an alternative ‘address’ domain to differentiate the antagonist binding mode of the MOR over the DOR and KOR. Therefore, a new compound containing specific structural features to interact with these amino acid residues might have increased selectivity for the MOR over the DOR and KOR.
Figure 2.
Naltrexone in MOR-binding pocket: mu opioid receptor model: ribbon and in cyan color; the residues in mu opioid receptor: ball and stick and in atom color; naltrexone molecular: ball and stick and carbon in yellow color, and oxygen red, nitrogen blue.
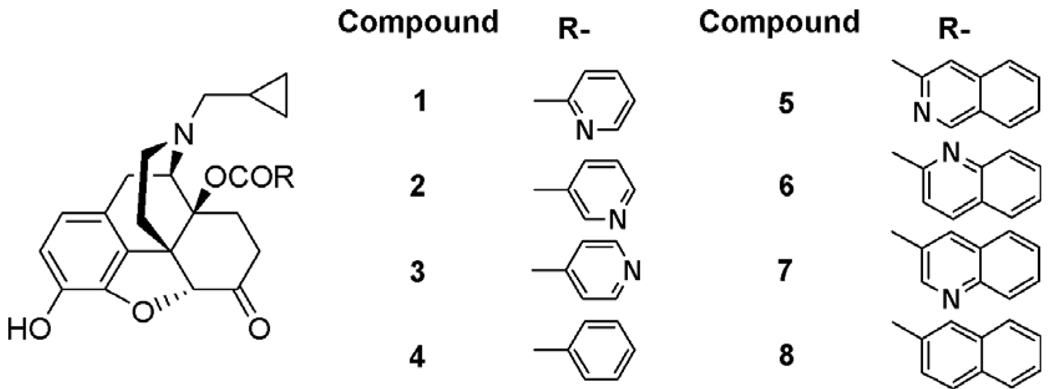
Based on this hypothesis, a series of novel 14-O-substituted naltrexone derivatives (Fig. 3) have been designed and synthesized. The ester bond in these novel ligands was assumed to provide a flexible conformation for the whole side chain. The nitrogen atom in the hetero-aromatic moiety on the 14-O-position of naltrexone was introduced to provide an opportunity for hydrogen bonding and/or aromatic stacking interaction with the amino acid residues Tyr212 and Trp320 in the MOR binding pocket (compounds 1–3 and 5–7). Compounds 4 and 8 were designed as control compounds to test this hypothesis. These ligands could also be considered as derivatives of clocinnamox without the Michael acceptor character.
Figure 3.
The designed ligand for primary study.
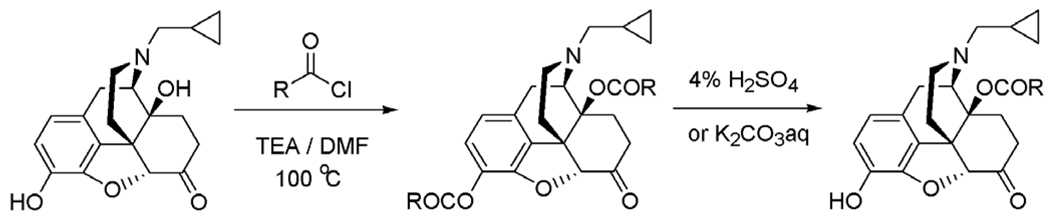
Using naltrexone as the starting material, the syntheses of these 14-O-heterocyclic-substituted derivatives was straightforward (Scheme 1). To be noticed, in the second step of the synthesis route, K2CO3 aqueous solution was used to prepare the control compounds 4 and 8 instead of using the acidic condition. All the final compounds were obtained with reasonable yield and characterized with NMR, IR, MS, and HPLC (see Supplementary information).
Scheme 1.
The synthetic route for the 14-O-subsituted naltrexone derivatives.
The primary biological studies of these ligands included competitive radioligand-binding assays using mono-cloned opioid receptors expressed in CHO cell lines. [3H] DAMGO, [3H] NTI and [3H] norBNI were used to label the MOR, DOR, and KOR, respectively. The binding affinities of these ligands for the MOR, DOR, and KOR, and comparative selectivities were summarized in Table 1. These compounds showed binding affinities in the subnanomolar-to-nanomolar range for the MOR.
Table 1.
Binding affinity and functional assay results for the 14-O-substituted naltrexone derivatives
| Compound |
Ki ± SEM (nM) |
Selectivity |
Percent Max of DAMGO | |||
|---|---|---|---|---|---|---|
| [3H]DAMGO (μ) | [3H] NTI (δ) | [3H] norBNI (κ) | δ/μ | κ/μ | ||
| Naltrexone | 0.26 ± 0.02 | 117.00 ± 8.90 | 5.15 ± 0.26 | 450 | 20 | 0.00 |
| β-FNA | 0.41 ± 0.04 | 27.78 ± 4.60 | 0.94 ± 0.05 | 68 | 2 | 0.00 |
| CTAP | 2.02 ± 0.71 | 1441.00 ± 106.10 | 1012.70 ± 174.80 | 713 | 501 | 0.00 |
| 1 | 0.14 ± 0.03 | 117.38 ± 17.97 | 25.50 ± 6.50 | 838 | 182 | 0.00 |
| 2 | 1.59 ± 0.61 | 170.30 ± 12.64 | 47.81 ± 8.48 | 107 | 30 | 0.00 |
| 3 | 5.58 ± 1.34 | 405.32 ± 234.68 | 49.21 ± 20.37 | 73 | 9 | 0.00 |
| 4 | 123.23 ± 38.23 | >10,000.00 | 586.42 ± 32.39 | >81 | 5 | 0.00 |
| 5 | 68.40 ± 6.04 | >10,000.00 | >10,000.00 | >146 | >146 | 0.00 |
| 6 | 1.44 ± 0.32 | 22.81 ± 19.52 | 67.15 ± 36.72 | 16 | 47 | 0.00 |
| 7 | 2.69 ± 0.72 | 818.43 ± 507.23 | 148.23 ± 55.53 | 304 | 55 | 22.00 ± 10.30 |
| 8 | 225.27 ± 46.6 | 907.18 ± 192.99 | 46.57 ± 13.53 | 4 | <1 | 0.00 |
The Ki values for the mu, delta and kappa opioid receptors were n = 3. The averages were reported along with their standard error of the means, SEM, for each compound. The comparison to percent stimulation of DAMGO was the Emax of the compound compared to the Emax of DAMGO (normalized to 100%). The DAMGO EC50 value was 45.1 ± 6.63 nM and its Emax value was 366 ± 23% stimulation over basal using a [35S]GTPγS functional assay. Naltrexone, β-FNA and CTAP were tested along as positive controls under the same conditions.
Also as shown above, all of these compounds exhibited different levels of selectivity for the MOR over the KOR and DOR. Among these, compound 1 had approximately 800-fold selectivity for the MOR over the DOR and nearly 200-fold selectivity over the KOR. Compound 5 also showed over 100-fold selectivity for the MOR over the other two receptor types, although its binding affinity for the MOR was significantly lower than compound 1. In addition, all of these compounds acted as MOR antagonists in 35[S]GTPγS functional assays except for compound 7, which was a partial agonist.
Compared to the control compounds 4 and 8, the MOR selectivity over DOR and KOR had been enhanced greatly in all of the other compounds. This result suggested that the 14-O-substitutions introduced onto the naltrexone skeleton might interact with the proposed alternative ‘address’ domain in the MOR, and the nitrogen atom in the heterocyclic ring might act as a hydrogen bond acceptor and play an important role for the selectivity. Among all of these ligands, compound 1 showed the highest selectivity, which suggested that it had the most favorable orientation of its side chain towards this plausible ‘address’-binding domain in the MOR. For compound 5, its side chain might confer selectivity for the MOR, whereas the bulkiness of its side chain also might have reduced its binding affinity for the MOR.
To further characterize compound 1 as the lead for our next generation molecular design, its antagonism was evaluated against DAMGO in 35[S]GTPγS functional assay. The concentration of compound 1 was 1.5 nM while DAMGO was in the range of 10–10,000 nM. The Ke value of compound was 0.20 ± 0.04 nM and apparent pA2 value was 9.72 ± 0.10. This observation was consistent with the binding affinity results and further verified that compound 1 could be used as the lead for future molecular design.
It has been reported by Schmidhammer et al., that 14-alkoxymorphinans showed very high opioid receptor affinity. These compounds exhibited significantly increased binding affinities at all opioid receptors without any specific preference for any one receptor type.41–43 Recently, Husbands et al., investigated the SAR of the analogs of clocinnamox, 14-aminodihydromorphinones, and 14-aminodihydrocodeinones, to explore the effect of changing the chain linking and substitution in the aromatic ring of cinnamoylaminomorphinones and codeinones.44–46 These authors found that a modest selectivity for the MOR over the DOR and KOR was achieved when the side chain on the 14 positions was comparably rotatable in these 14-aminiodihydromorphinone compounds.
Comparing to the compounds reported by Schmidhammer and Husbands, the compounds reported here showed similar affinity for the MOR, but much higher selectivity over the DOR and KOR. One possible explanation might be that the introduction of a shorter side chain and a more flexible ester bond in our compounds might lead to a more favorable conformation and orientation of the side chain to target the ‘address’ locus and thereby improve selectivity for the MOR. Certainly this ‘address’ locus needs to be further verified, for example, by site-directed mutagenesis, in future studies.
In summary, a series of 14-O-heterocyclic-substituted naltrexone derivatives were designed, synthesized, and evaluated as selective MOR antagonists. Most of these novel ligands exhibited subnanomolar-to-nanomolar binding affinity for the MOR, with compound 1 showing the highest selectivity for the MOR over the DOR and KOR. These results implicated a plausible ‘address’ domain in the extracellular loops of the MOR. The knowledge gained from these studies will enrich the ‘message-address’ concept that has been applied successfully in opioid research and may lead to the identification of potent MOR-selective non-peptide antagonists.
Supplementary Material
Acknowledgments
We thank Dr. Lee-Yuan Liu-Chen at Temple University and Dr. Ping-Yee Law at the University of Minnesota for the generous gift of opioid receptor expressing CHO cell lines. We appreciate the generous help from Mallinckrodt, Inc. for the gift of naltrexone sample. The work was partially supported by PHS Grant DA10770 from NIH/NIDA(D.E.S.), the Research Fund 646920 from A.D. Williams Foundation and PHS Grant DA24022 from NIH/NIDA (Y.Z.).
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2008.12.093.
References and notes
- 1.Goldstein A, Naidu A. Mol. Pharmacol. 1989;36:265. [PubMed] [Google Scholar]
- 2.Dhawan BN, Cesselin F, Raghubir R, Reisin T, Bradley PB, Portoghese PS, Hamon M. Pharmacol. Rev. 1996;48:567. [PubMed] [Google Scholar]
- 3.Minami M, Satoh M. Neurosci. Res. 1995;23:121. doi: 10.1016/0168-0102(95)00933-k. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman DM, Leander JD. J. Med. Chem. 1990;33:895. doi: 10.1021/jm00165a002. [DOI] [PubMed] [Google Scholar]
- 5.Schmidhammer H. Prog. Med. Chem. 1998;35:83. [PubMed] [Google Scholar]
- 6.Eguchi M. Med. Res. Rev. 2004;242:182. doi: 10.1002/med.10059. [DOI] [PubMed] [Google Scholar]
- 7.Skoubis PD, Matthes HW, Walwyn WM, Kieffer BL, Maidment NT. Neuroscience. 2001;106:757. doi: 10.1016/s0306-4522(01)00333-5. [DOI] [PubMed] [Google Scholar]
- 8.Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dollé P, Tzavara E, Hanoune J, Roques BP, Kieffer BL. Nature. 1996;383:819. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 9.Gold MS, Dackis CA, Pottash AL, Sternbach HH, Annitto WJ, Martin D, Dackis MP. Med. Res. Rev. 1982;23:211. doi: 10.1002/med.2610020302. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez JP, Brogden RN. Drugs. 1988;35:192. doi: 10.2165/00003495-198835030-00002. [DOI] [PubMed] [Google Scholar]
- 11.Goodman AJ, Le Bourdonnec B, Dolle RE. ChemMedChem. 2007;2:1552. doi: 10.1002/cmdc.200700143. [DOI] [PubMed] [Google Scholar]
- 12.Schwyzer R. Ann. N.Y. Acad. Sci. 1977;297:3. doi: 10.1111/j.1749-6632.1977.tb41843.x. [DOI] [PubMed] [Google Scholar]
- 13.Portoghese PS, Lipkowski AW, Takemori AE. Life Sci. 1987;40:1287. doi: 10.1016/0024-3205(87)90585-6. [DOI] [PubMed] [Google Scholar]
- 14.Jones RM, Hjorth SA, Schwartz TW, Portoghese PS. J. Med. Chem. 1998;41:4911. doi: 10.1021/jm9805182. [DOI] [PubMed] [Google Scholar]
- 15.Portoghese PS, Sultana M, Nagase H, Takemori AE. J. Med. Chem. 1988;31:281. doi: 10.1021/jm00397a001. [DOI] [PubMed] [Google Scholar]
- 16.(a) Schmidhammer H, Burkard WP, Eggstin-Aeppli L, Smith CFC. J. Med. Chem. 1989;32:418. doi: 10.1021/jm00122a021. [DOI] [PubMed] [Google Scholar]; (b) Schmidhammer H, Smith CF, Erlach D, Koch M, Krassnig R, Schwetz W, Wechner C. J. Med. Chem. 1990;33:1200. doi: 10.1021/jm00166a018. [DOI] [PubMed] [Google Scholar]; (c) Schmidhammer H, Smith CF, Erlach D, Koch M, Krassnig R, Schwetz W, Wechner C. Prog. Clin. Biol. Res. 1990;328:37. [PubMed] [Google Scholar]
- 17.(a) Lewis JW, Smith CFC, McCarthy PS, Kobylecki RJ, Myers M, Haynes AS, Lewis CJ, Waltham K. NIDA Res. Monogr. 1988;90:136. [PubMed] [Google Scholar]; (b) Portoghese PS, Takemori AE. NIDA Res. Monogr. 1986;69:157. [PubMed] [Google Scholar]; (c) Burke TF, Woods JH, Lewis JW, Medzihradsky F. J. Pharmacol. Exp. Ther. 1994;2712:715. [PubMed] [Google Scholar]
- 18.(a) Xue J-C, Chen C, Zhu J, Kunapuli SP, De Riel JK, Yu L, Liu-Chen L-Y. J. Biol. Chem. 1995;27022:12977. [PubMed] [Google Scholar]; (b) Zhu J, Xue J-C, Law P-Y, Claude PA, Luo L-Y, Yin J, Chen C, Liu-Chen L-Y. FEBS Lett. 1996;384:198. doi: 10.1016/0014-5793(96)00312-2. [DOI] [PubMed] [Google Scholar]
- 19.(a) Bonner G, Meng F, Akil H. Eur. J. Pharmcol. 2000;403:37. doi: 10.1016/s0014-2999(00)00578-1. [DOI] [PubMed] [Google Scholar]; (b) Xu H, Lu YF, Partilla JS, Zheng QX, Wang JB, Brine GA, Carroll FI, Rice KC, Chen KX, Chi ZQ, Rothman RB. Synapse (New York) 1999;32:23. doi: 10.1002/(SICI)1098-2396(199904)32:1<23::AID-SYN3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Okada T, Le Trong I, Fox BA, Behnke CA, Stenkamp RE, Palczewski K. J. Struct. Biol. 2000;130:73. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 21.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Biochemistry. 2001;40:7761. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salom D, Le Trong I, Pohl E, Ballesteros JA, Stenkamp RE, Palczewski K, Lodowski DT. J. Struct. Biol. 2006;156:497. doi: 10.1016/j.jsb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16123. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Nature. 2007;450:383. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 27.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Nature. 2008 June 25; doi: 10.1038/nature07101. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patny A, Desai PV, Avery MA. Curr. Med. Chem. 2006;13:1667. doi: 10.2174/092986706777442002. [DOI] [PubMed] [Google Scholar]
- 31.Ballesteros JA, Shi L, Javitch JA. Mol. Pharmacol. 2001;60:1. [PubMed] [Google Scholar]
- 32.Becker OM, Shacham S, Marantz Y, Noiman S. Curr. Opin. Drug. Discov. Dev. 2003;63:353. [PubMed] [Google Scholar]
- 33.Moro S, Spalluto G, Jacobson KA. Trends Pharmacol. Sci. 2005;261:44. doi: 10.1016/j.tips.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Nowak M, Kolaczkowski M, Pawlowski M, Bojarski AJ. J. Med. Chem. 2006;491:205. doi: 10.1021/jm050826h. [DOI] [PubMed] [Google Scholar]
- 35.McLean TH, Chambers JJ, Parrish JC, Braden MR, Marona-Lewicka D, Kurrasch-Orbaugh D, Nichols DE. J. Med. Chem. 2006;4914:4269. doi: 10.1021/jm060272y. [DOI] [PubMed] [Google Scholar]
- 36.Hobrath JV, Wang S. J. Med. Chem. 2006;4915:4470. doi: 10.1021/jm0501634. [DOI] [PubMed] [Google Scholar]
- 37.Singh S, Malik BK, Sharma DK. Chem. Biol. Drug. Des. 2007;693:191. doi: 10.1111/j.1747-0285.2007.00478.x. [DOI] [PubMed] [Google Scholar]
- 38.Patny A, Desai PV, Avery MA. Proteins. 2006;654:824. doi: 10.1002/prot.21196. [DOI] [PubMed] [Google Scholar]
- 39.Lu ZL, Saldanha JW, Hulme EC. Trends Pharmacol. Sci. 2002;233:140. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Sham YY, Rajamani R, Gao JL, Portoghese PS. ChemBioChem. 2005;6:859. doi: 10.1002/cbic.200400207. [DOI] [PubMed] [Google Scholar]
- 41.Lattanzi R, Spetea M, Schullner F, Rief SB, Krassnig R, Negri L, Schmidhammer H. J. Med. Chem. 2005;48:3372. doi: 10.1021/jm040894o. [DOI] [PubMed] [Google Scholar]
- 42.Spetea M, Schüllne F, Moisa RC, Berzetei-Gurske IP, Schraml B, Dorfler C, Aceto MD, Harris LS, Coop A, Schmidhammer H. J. Med. Chem. 2004;47:3242. doi: 10.1021/jm031126k. [DOI] [PubMed] [Google Scholar]
- 43.Greiner E, Spetea M, Krassnig R, Schullne F, Aceto M, Harris LS, Traynor JR, Woods JH, Coop A, Schmidhammer H. J. Med. Chem. 2003;46:1758. doi: 10.1021/jm021118o. [DOI] [PubMed] [Google Scholar]
- 44.Rennison D, Moynihan H, Traynor JR, Lewis JW, Husbands SM. J. Med. Chem. 2006;49:6104. doi: 10.1021/jm060595u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieland NP, Moynihan HA, Carrington S, Broadbear J, Woods JH, Traynor JR, Husbands SM, Lewis JW. J. Med. Chem. 2006;4917:5333. doi: 10.1021/jm0604777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grundt P, Jales AR, Traynor JR, Lewis JW, Husbands SM. J. Med. Chem. 2003;46:1563. doi: 10.1021/jm021073r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.