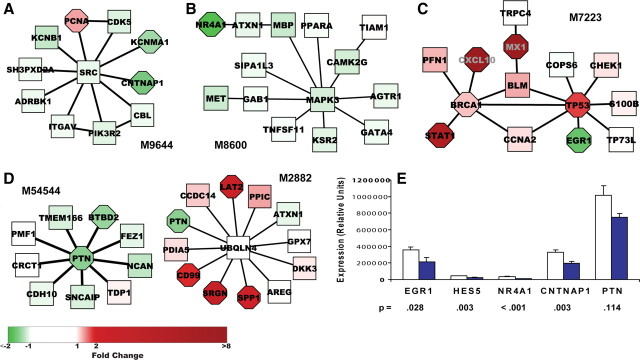
Figure 3.
Significant modules containing selected downregulated genes and RT-PCR confirmation. A–D, Significant modules in which downregulated genes appear. Module M9644 (Fig. 3A) was the highest scoring downregulated module and is significantly enriched for genes participating in signal transduction (PCNA, ITGAV, ADRBK1, CDK5, CBL, CNTNAP1, PIK3R2, SRC). Fold change in gene expression relative to uninfected monkeys is depicted by intensity of node color: red (upregulated) or green (downregulated). Significance of differential expression as determined by Cyber T p value is represented by node shape: rectangular (p > 10−3), octagonal (≤10−3). E, Hippocampal tissue from each of the nine uninfected and nine SIVE monkeys was tested by RT-PCR for four downregulated genes present in significant modules and HES5. Normalization was done using housekeeping genes 18S, TBP, and GAPDH. Figure shows mean expression ± SEM in terms of relative units; t test was performed on log-transformed data. Unfilled bars show control data; solid blue bars show SIVE data.