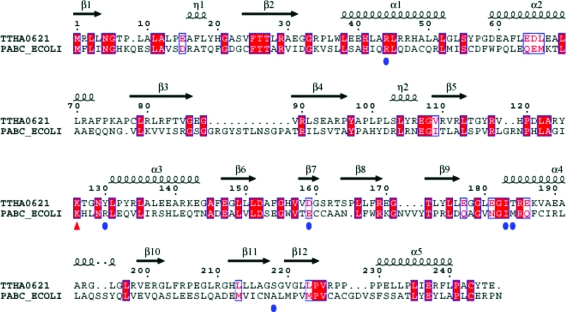
Figure 3.
Sequence alignment of the putative 4-amino-4-deoxychorismate lyase (TTHA0621) from T. thermophilus (Q5SKM2_THET8) and the 4-amino-4-deoxychorismate lyase (PabC) from E. coli (PABC_ECOLI). The secondary-structural features of TTHA0621 are indicated above the alignment. The residues that potentially interact with the PLP cofactor are indicated by blue circles. The conserved catalytic residue Lys161, which forms a Schiff base with PLP, is indicated by a red triangle. The colours reflect the similarity (conserved residues are shown as white characters on a red background, residues that are similar within a group are shown as red characters and residues that are similar across groups are shown in blue frames). 310-Helices are denoted by the symbol η. The sequence was aligned and displayed using ClustalW (Thompson et al., 1994 ▶) and ESPript (Gouet et al., 1999 ▶), respectively.