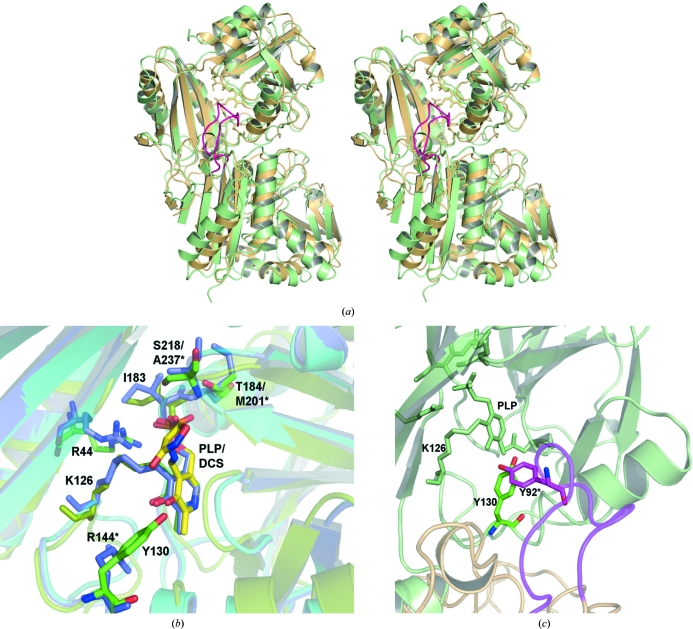
Figure 4.
Comparison of TTHA0621 with E. coli PabC. (a) Superposition of TTHA0621 (light orange) on E. coli PabC (light green) for the Cα atoms corresponding to one subunit (top) of the dimer (PDB code 1i2k; Parsons et al., 2002 ▶). In the E. coli PabC dimer a long flexible loop (pink) from one monomer, which is inserted between strands β3 and β4, protrudes into the catalytic site of the other monomer. (b) Near the cofactor-binding site region. The interacting residues and the PLP cofactors of the THA0621 and E. coli PabC complexes are shown in green and slate, respectively. The interacting residues and the inhibitor DCS in the E. coli PabC–DCS complex are shown in yellow. (c) Near the O3′ atom of PLP, conservation of Tyr is observed between the T. thermophilus and E. coli enzymes. In the E. coli PabC structure Tyr92, which lies in the flexible region (pink) of the neighbouring subunit, forms intermolecular interactions with the PLP cofactor bound to the other subunit, whereas in TTHA0621 a water molecule bridges Tyr130 and PLP within the monomer. The nonconserved residues in E. coli PabC are labelled with asterisks.