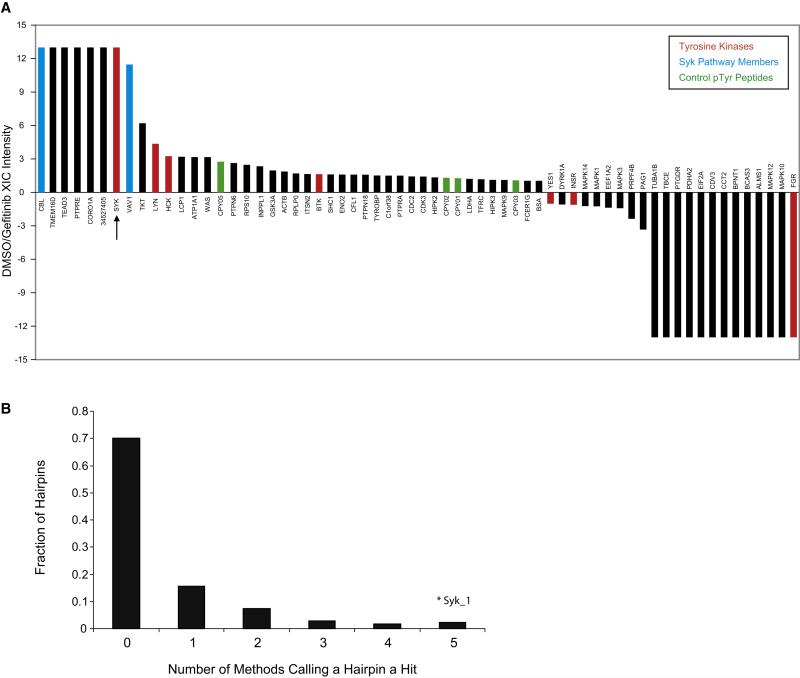
Figure 1. Proteomic and RNAi-based approaches identify Syk as a target for AML differentiation.
(A) Peptide-IP/LC-MS/MS was used to identify proteins dephosphorylated upon gefitinib treatment. HL-60 cells were treated with vehicle (DMSO) or gefitinib at 10 μM for 10 minutes. Phosphotyrosine peptides were identified in Syk, Cbl, and Vav1 that were dephosphorylated with gefitinib. The LC-MS/MS extracted ion currents (XIC) for all identified pTyr peptide precursor ions belonging to an individual protein were summed and used to calculate differential expression ratios in DMSO- and gefitinib-treated cells. The specific phosphopeptides and pTyr sites observed are shown in Table S1. CPY01,2,3,5 are control pTyr peptides spiked into each sample at the same concentration. Positive ratios indicate higher expression for DMSO treatment, negative ratios indicate higher expression for gefitinib treatment. Ratios of 13 and −13 indicate that phosphopeptides for a protein were only detected in one of the treatments.
(B) High-throughput RNAi Screen Performance. Five scoring methods were used to identify hairpins inducing a myeloid differentiation signature (Summed Score, Weighted Summed Score, Naïve Bayes, K-Nearest Neighbor, and Support Vector Machine). The majority of shRNAs did not score by any method. The SYK-specific shRNA (Syk_1) scored in all methods.