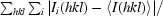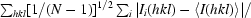Table 1. Summary of 2.0 Å lysozyme data sets.
Values in parentheses are for lowest resolution (∞–6.32 Å) and highest resolution (2.11–2.00 Å) bins. The actual low-resolution limit of the data was 15–20 Å.
| Name | Laue best | Laue firsts | Laue small firsts | Laue single-crystal | Mono complete | Mono small |
|---|---|---|---|---|---|---|
| Crystals/frames | 7/36 | 18/18 | 5/5 | 1/5 | 1/60 | 1/22 |
| Completeness (%) | 89.6 (83.3, 74.9) | 90.2 (86.9, 81.3) | 70.4 (61.3, 49.1) | 70.9 (52.7, 50.8) | 98.3 (89.2, 99.4) | 70.7 (54.0, 71.4) |
| Multiplicity | 7.7 (4.5, 4.1) | 5.5 (4.0, 3.0) | 2.2 (1.8, 1.6) | 2.3 (1.9, 1.6) | 4.5 (3.5, 4.6) | 2.3 (2.3, 2.3) |
| Rmerge† | 0.174 (0.098, 0.300) | 0.200 (0.150, 0.217) | 0.144 (0.125, 0.151) | 0.127 (0.069, 0.206) | 0.052 (0.046, 0.171) | 0.049 (0.032, 0.118) |
| Rp.i.m.‡ | 0.057 (0.048, 0.139) | 0.072 (0.074, 0.112) | 0.093 (0.099, 0.119) | 0.080 (0.051, 0.157) | 0.027 (0.024, 0.083) | 0.036 (0.023, 0.083) |
| MOLREP score | 0.524 | 0.528 | 0.485 | 0.494 | 0.551 | 0.494 |
| R after rigid-body refinement | 0.392 | 0.394 | 0.390 | 0.384 | 0.399 | 0.394 |
| R/Rfree after all refinement | 0.266/0.318 | 0.259/0.319 | 0.256/0.331 | 0.264/0.320 | 0.241/0.265 | 0.228/0.266 |