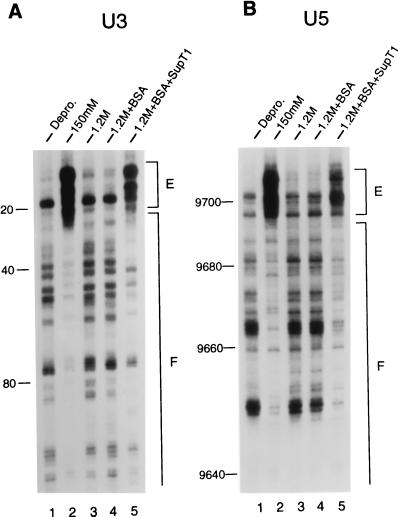
Figure 2.
Structural reconstitution of the HIV-1 intasome with uninfected cell extract. Mu transposition reactions were deproteinized and analyzed by denaturing sequencing gels after two rounds of PCR. The U3 end of HIV-1 DNA was analyzed in A, and the U5 end was analyzed in B. Lanes 1, the pattern of Mu transposition into deproteinized cDNA. Lanes 2, the native structure of the HIV-1 intasome. Note the regions of strong transpositional enhancements (E) and DNA footprints (F) relative to the deproteinized control. Lanes 3, salt-stripped PICs. Note the similarity in patterns between the salt-stripped and deproteinized PICs. Lanes 4, reconstitution buffer control. Lanes 5, salt-stripped PICs reconstituted with 5 μl of uninfected cell extract. The reaction products were run alongside DNA-sequencing ladders to determine the distance from the ends of HIV-1 (nucleotide position 1 in U3 and position 9,718 in U5). The U3 and U5 footprints extended approximately 250 and 200 bp, respectively, in the native structure.