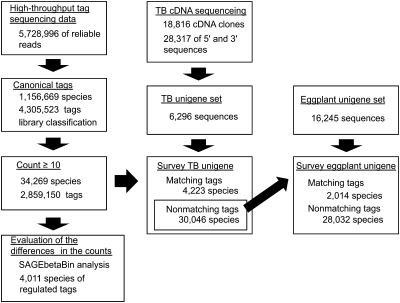
Fig. 2.
A flow chart for the analysis of the high-throughput sequencing data of expression tag libraries. Reliable sequence reads were checked if they had canonical ‘adaptor-tag’ features. Expression tag sequences of 24 bp were extracted and classified into four library categories according to the library coding nucleotides. The tag species that were counted ≥10 times in all four libraries were chosen for further analysis. For reference sequences required for the assignment of the tag sequences, TB cDNAs were intensively sequenced, and 28 317 cDNA sequences were assembled, resulting in 6296 TB unigene sequences. The eggplant unigene set comprising 16 245 sequences was also used as a reference. The TB unigene set was surveyed for the unigene sequences containing exact tag sequences. For 30 046 non-matching sequences, the eggplant unigene set was similarly searched. The differences in the counts of individual tag species between Cd-treated and untreated libraries were evaluated by the SAGEbetaBin analysis with a Bayes error rate of 0.05 (Vêncio et al., 2004).