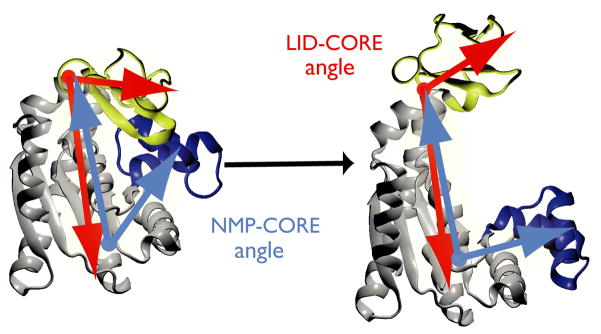
Figure 1. Structural transition of adenylate kinase (AdK).
The enzyme consists of three well-defined domains: the rigid, predominantly β-sheet ‘CORE’ (gray, AKeco residues 1–29, 60–121, 160–214), the nucleotide monophosphate binding domain (‘NMP’, blue, 30–59), and the ‘LID’ domain (yellow, residues 122–159). The NMP-CORE angle θNMP is formed by the centres of geometry of the backbone and Cβ atoms in residues 115–125 (CORE/LID), 90–100 (CORE), and 35–55 (NMP) of E. coli AdK. θLID is defined equivalently as the angle between 179–185 (CORE), 115–125 (CORE/hinge/LID), and 125–153 (LID).