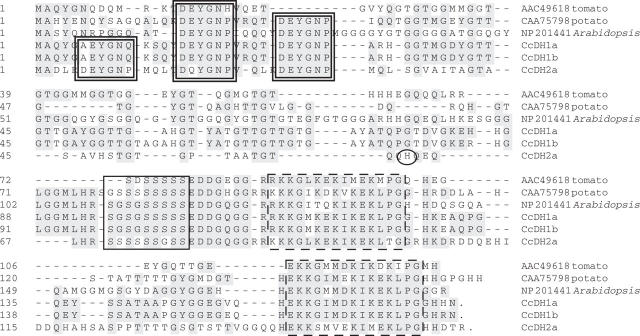
Fig. 1.
The optimized alignment of C. canephora Y3SK2-type dehydrins with several of the closest plant homologues. This alignment was generated using the Clustal W program in the MegAlign software (DNASTAR) and then further optimized manually. Shaded blocks indicate identical amino acids. The doubled rectangles demarcate the Y-segments, the single dark rectangles demarcate the S-segments, and the rectangles with broken lines demarcate the K-segments. The black circle represents the single amino acid difference between CcDH2a and CcDH2b. Accession numbers: CcDH1a, DQ323987; CcDH1b, DQ323988; CCDH2a, DQ323989; Lycopersicon esculentum dehydrin TAS14, AAC49618; Solanum commersonii dehydrin Dhn1, CAA75798; Arabidopsis thaliana dehydrin RAB18, NP_201441.