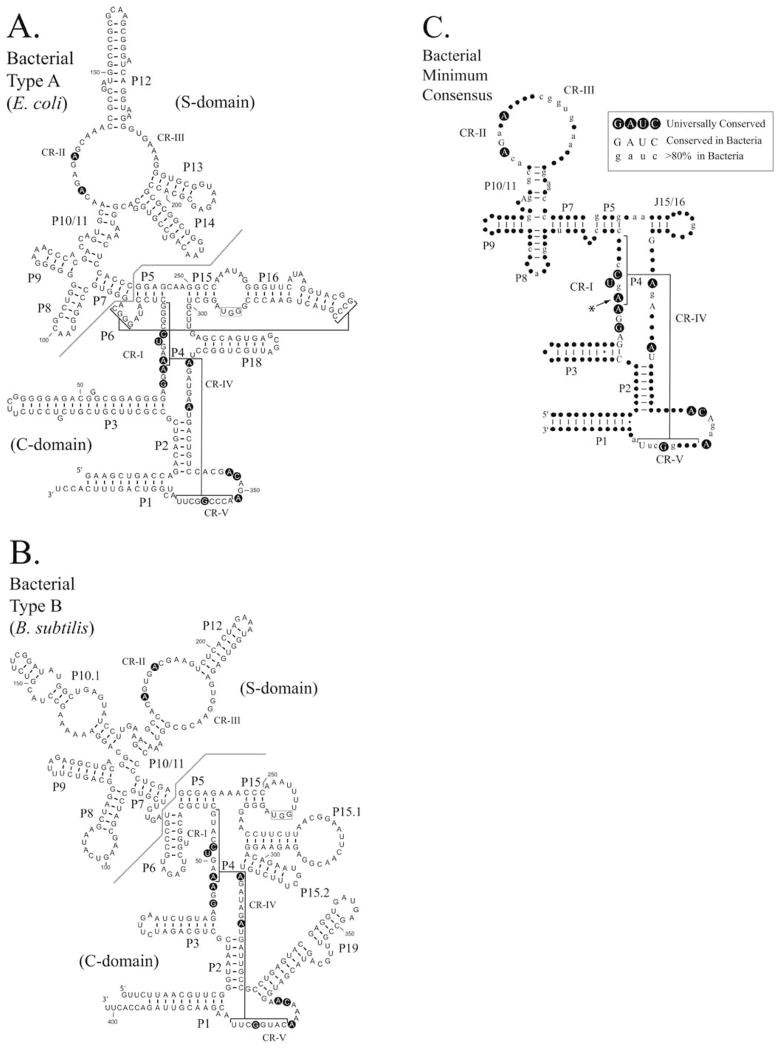
FIGURE 2.
Comparison of Type A and Type B bacterial RNase P RNAs. The secondary structures of RNase P RNAs derived by phylogenetic comparative sequence analysis are shown (Haas and Brown, 1998). (A). The secondary structure of the E. coli RNase P RNA (Type A). (B). The secondary structure of the B. subtilis RNase P RNA (Type B). Helices are numbered as paired regions (e.g. P1), other helices are indicated by lines (e.g. P4 and P6). Regions of the RNA implicated in substrate binding the “specificity domain” (S-domain) and catalytic activity the “catalytic domain” (C-domain) are separated by a line. A boxed region within the loop of P15 indicates a binding site for the 3′CCA of the pre-tRNA substrate, which is present in many bacterial RNase P RNAs. Nucleotides conserved in all known RNase P RNAs are shown in bold background. (C). Consensus structure showing elements common to all known bacterial RNase P RNAs (figure adapted from Marquez et al., 2005). The highlighted conserved adenine has been shown to be extrahelical in the structure of the Bacillus stearothermophilus RNase P RNA (Kazantsev et al., 2005). In this structure, the adjacent conserved adenosine forms the corresponding base pair within the P4 helix.