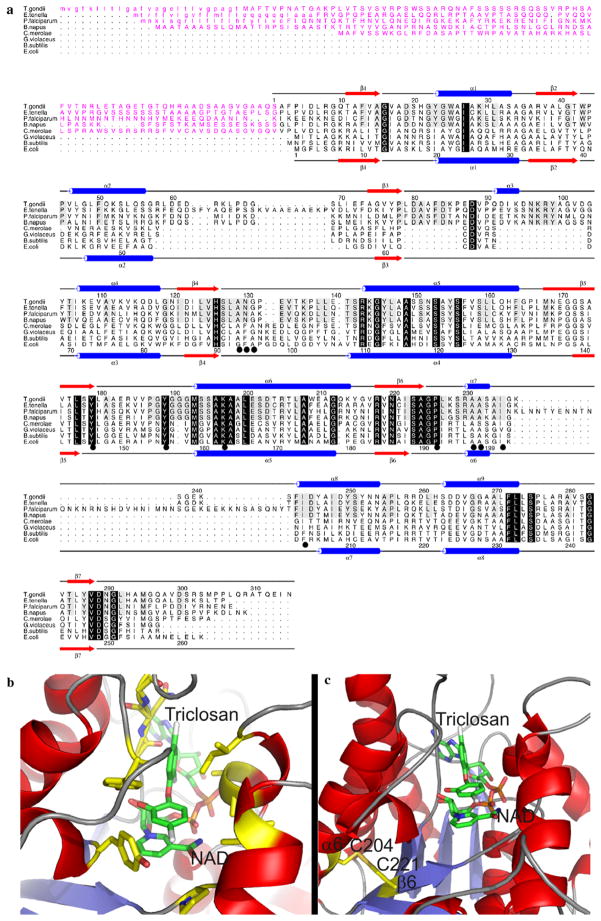
Fig. 1.
(a) A structure-based sequence alignment of ENRs from Toxoplasma gondii, Eimeria tenella, Plasmodium falciparum, Brassica napus, Gloeobacter violaceus, Cyanidioschyzon merolae, Bacillus subtilis and Escherichia coli. The secondary structure elements and sequence numbering for T. gondii and E. coli ENR are shown on the top and bottom of the alignment, respectively, with α-helices displayed as blue cylinders and β-sheets as red arrows. Those residues which are conserved across the ENR family are highlighted by a black box with reverse type, with those residues conserved among the apicomplexans being boxed in grey. Those residues which have been shown to be involved in triclosan binding are underscored by a black circle. The transit peptides are shown in purple and the predicted cleavable von Heijne signal sequences are shown in lower case. (b) The structure of T. gondii ENR onto which the residues for E. tenella ENR were mutated (in silico) based on the structural alignment in A with those residues involved in triclosan binding (which are fully conserved between T. gondii ENR and EtENR) shown in a stick format and highlighted by a black circle in the alignment in (a). (c) A structural representation of the position of two2 Cys residues on α6 and β6, which are within the appropriate distance for disulphide bond formation. For both (b) and (c) the stick representations are coloured red, blue, orange, gold and grey for oxygen, nitrogen, phosphorous, sulphur and chlorine, respectively, with carbon being coloured green for NAD+ and triclosan and yellow for all other residues.