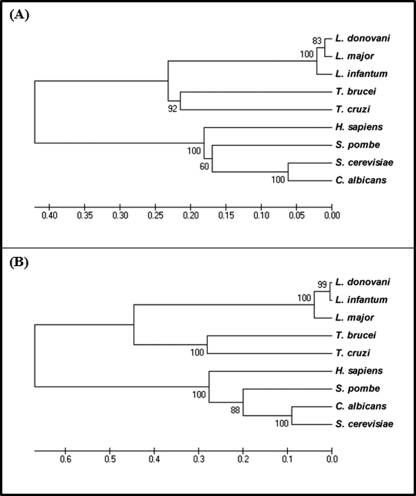
FIGURE 4.
A, phylogenetic analysis by Unweighted Pair Group Method with Arithmetic Mean of DHS-like gene, DHSL20. The phylogram presented was a consensus of 500 bootstrap replicates constructed using the MEGA program (version 4.0). The numbers at the node present the percentage of trees with the same node among all of the bootstraps. The accession numbers of the gene sequences used are as follows: L. donovani (ABP65295), L. major (LmjF20.0250), L. infantum (LinJ20.0270), T. brucei (Tb927.1.870), T. cruzi (Tc00.1047053511421.60), Homo sapiens (P49366), Schizosaccharomyces pombe (CAA22194), S. cerevisiae (P38791), Candida albicans (XM_716612.1). B, phylogenetic analysis by Unweighted Pair Group Method with Arithmetic Mean of the DHS34 gene. The phylogram presented was a consensus of 500 bootstrap replicates constructed using the MEGA program (version 4.0). The numbers at the node present the percentage of trees with the same node among all the bootstraps. The accession numbers of the gene sequences used are as follows: L. donovani (ACF75531), L. major (LmjF34.0330), L. infantum (LinJ34.0350), T. brucei (Tb10.70.4900), T. cruzi (Tc00.1047053504119.29), H. sapiens (P49366), S. pombe (NP_595146.1), C. albicans (XP_721705.1), S. cerevisiae (P38791).