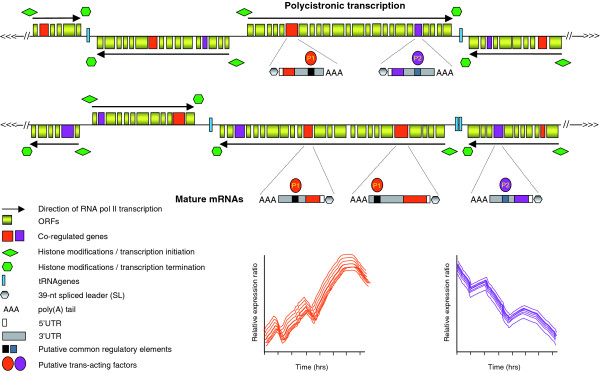
Figure 1.
Coordinated post-transcriptional regulation of T. brucei mRNAs during differentiation. Schematic diagram of putative regions of two T. brucei chromosomes. Genes in T. brucei are organized into long polycistronic clusters that are co-transcribed by RNA polymerase II (Pol II) to yield polycistronic pre-mRNAs, which are processed by trans-splicing (addition of a capped spliced leader RNA of 39 nucleotides to the 5' terminus of transcripts) and 3'-polyadenylation to generate mature mRNAs. Transcription initiates from divergent strand-switch regions (SSRs) and terminates at convergent SSRs, where tRNA genes are often located (although they can be present at non-SSRs). Initiation and termination of transcription in T. brucei are characterized by distinct chromatin variants and modifications [9]. Three recent reports [1-3] indicate that subsets of trypanosome genes form post-transcriptional regulons during T. brucei life-cycle transitions. Two hypothetical post-transcriptional regulons formed during differentiation are shown. Subsets of genes (here shown in either orange or violet) have common regulatory elements or conserved secondary structures within the 3'UTRs. These are recognized by trans-acting factors (specific for either the set of genes in orange or in violet, and either stabilizing or destabilizing mRNAs), which allow a coordinated regulation of sets of mRNAs. This is illustrated in the two lower graphs, where mRNA levels are plotted against the differentiation process with time. The mRNA levels of the cluster of genes in orange are increasing coordinately upon differentiation, whereas the cluster of genes in violet are decreasing upon differentiation in a coordinated fashion.