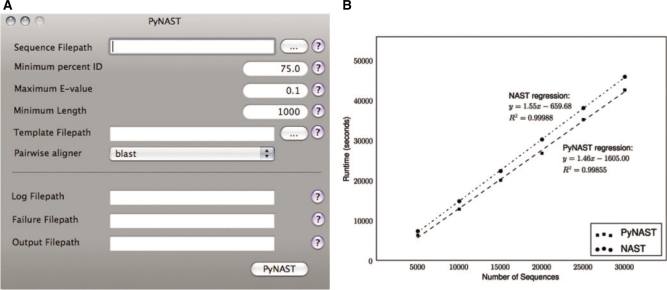
Fig. 1.
(A) Screenshot of the PyNAST graphical user interface for Mac OS X. (B) Runtime of PyNAST is compared with that of NAST, each running on a single processor. PyNAST has a slightly shorter per sequence runtime (slope). The candidate sequences used in this evaluation ranged from 917 to 1343 bases, with a median length of 1294. The template alignment was a Greengenes core set (dated November 8, 2007) with 7682 positions and 4938 sequences.