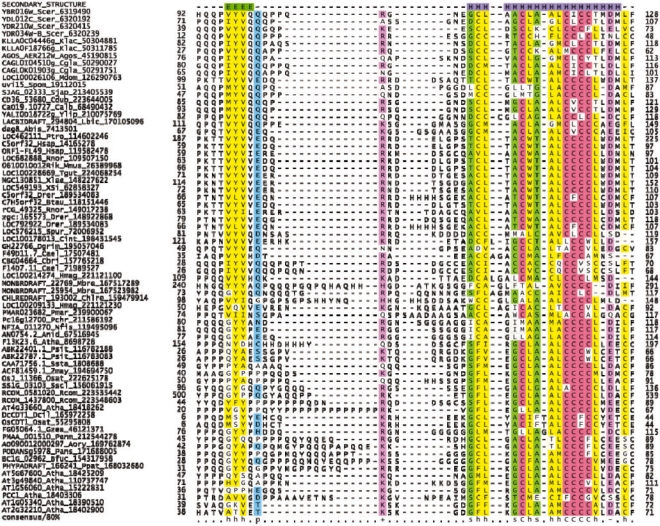
Fig. 1.
Multiple sequence alignment of the CYSTM domain superfamily. The columns were colored according to the consensus shown below the alignment, and the predicted secondary structure is shown on top. The sequences are labeled using the gene name, species abbreviation and GenBank gi number and sequence identifiers. The species abbreviations are—Abis: Agaricus bisporus; Atha: Arabidopsis thaliana; Agos: Ashbya gossypii; Anid: Aspergillus nidulans; Aory: Aspergillus oryzae; Btau: Bos taurus; Bfuc: Botryotinia fuckeliana; Cbri: Caenorhabditis briggsae; Cele: Caenorhabditis elegans; Calb: Candida albicans; Cdub: Candida dubliniensis; Cgla: Candida glabrata; Crei: Chlamydomonas reinhardtii; Cint: Ciona intestinalis; Drer: Danio rerio; Dcil: D.ciliaris; Dgri: Drosophila grimshawi; Gzea: Gibberella zeae; Hsap: Homo sapiens; Hmag: Hydra magnipapillata; Klac: Kluyveromyces lactis; Lbic: Laccaria bicolor; Mdom: Monodelphis domestica; Mbre: Monosiga brevicollis MX1; Mmus: Mus musculus; Nfis: Neosartorya fischeri; Osat: O.sativa; Ptro: Pan troglodytes; Pchr: Penicillium chrysogenum; Pmar: Penicillium marneffei; Pmar: Perkinsus marinus; Ppat: Physcomitrella patens; Psit: Picea sitchensis; Pans: Podospora anserina; Rnor: Rattus norvegicus; Rcom: Ricinus communis; Scer: S.cerevisiae; Sjap: Schizosaccharomyces japonicus; Spom: S.pombe; Sscl: Sclerotinia sclerotiorum; Ssta: Sporobolus stapfianus; Spur: Strongylocentrotus purpuratus; Tgut: Taeniopygia guttata; Xtro: Xenopus tropicalis; Xlae: Xenopus laevis; Ylip: Yarrowia lipolytica; Zmay: Zea mays.