Abstract
The amyloid precursor protein (APP) is a ubiquitously expressed transmembrane adhesion protein and the progenitor of amyloid-β peptides. The major splice isoforms of APP expressed by most tissues contain a Kunitz protease inhibitor domain; secreted APP containing this domain is also known as protease nexin 2 and potently inhibits serine proteases, including trypsin and coagulation factors. The atypical human trypsin isoform mesotrypsin is resistant to inhibition by most protein protease inhibitors and cleaves some inhibitors at a substantially accelerated rate. Here, in a proteomic screen to identify potential physiological substrates of mesotrypsin, we find that APP/protease nexin 2 is selectively cleaved by mesotrypsin within the Kunitz protease inhibitor domain. In studies employing the recombinant Kunitz domain of APP (APPI), we show that mesotrypsin cleaves selectively at the Arg15-Ala16 reactive site bond, with kinetic constants approaching those of other proteases toward highly specific protein substrates. Finally, we show that cleavage of APPI compromises its inhibition of other serine proteases, including cationic trypsin and factor XIa, by 2 orders of magnitude. Because APP/protease nexin 2 and mesotrypsin are coexpressed in a number of tissues, we suggest that processing by mesotrypsin may ablate the protease inhibitory function of APP/protease nexin 2 in vivo and may also modulate other activities of APP/protease nexin 2 that involve the Kunitz domain.
Keywords: Blood Coagulation/Coagulation Factors, Blood Coagulation/Serpins, Enzymes/Peptidases, Enzymes/Proteolytic, Protease, Protease/Inhibitor, Proteases/Serine protease
Introduction
Mesotrypsin is a human trypsin encoded by the PRSS3 gene found on chromosome 9p13 (1). Normal expression of PRSS3 is restricted to pancreas, brain, and, to a lesser extent, small intestine and colon (2–4); additionally, PRSS3 appears to be transcriptionally up-regulated with cancer progression in epithelial cancers, including lung (5), colon (6), and prostate.2 Mesotrypsin exhibits substantially different specificity from other trypsins toward protein substrates. It fails to activate pancreatic zymogens and also shows reduced capacity to degrade trypsinogens (7). Compared with other trypsins, it is significantly compromised in its ability to cleave protease-activated receptors (8–10). Despite limited activity toward these classic trypsin substrates, mesotrypsin displays enhanced catalytic activity compared with other trypsins in the cleavage of certain specific protein substrates, most notably several canonical protease inhibitors (7, 11).
The “canonical” inhibitors of serine proteases, named for a protease-binding loop of highly characteristic backbone conformation (12, 13), fulfill the paradoxical function of binding to a protease in a substrate-like manner yet acting as an inhibitor rather than an ordinary substrate. These inhibitors, representing at least 18 different convergently evolved protein families (14, 15), inhibit their cognate proteases via the “Laskowski mechanism,” in which inhibitors act as highly specific, limited proteolysis substrates for target enzymes (14, 16). They bind so as to position a specific peptide bond, the “reactive site,” in the active site of the enzyme, ideally oriented for enzymatic cleavage (17), but due to a binding affinity many orders of magnitude stronger than that associated with ordinary substrate binding and to interactions at the protease-inhibitor interface that deter progress of the reaction and dissociation of the cleaved product peptides, enzymatic turnover proceeds extremely slowly. The human genome encodes more than 50 known or putative canonical inhibitors belonging to three families: the I1 or Kazal family, the I2 or Kunitz family, and the I17 or WAP family (MEROPS data base, available on the World Wide Web) (18). Some of these inhibitors are very small, single domain proteins, whereas others represent functional protease inhibitor domains within larger proteins.
The amyloid precursor protein (APP)3 is a transmembrane protein present in the plasma membranes of all human cell types. Alternative splicing yields three major isoforms of APP: the ubiquitously expressed isoforms APP751 and APP770 contain a 56-residue canonical inhibitor domain belonging to the Kunitz family, whereas the shorter APP695 lacks the Kunitz inhibitor domain and is primarily expressed in the nervous system (19–21). As suggested by its name, APP is the precursor of the amyloidogenic peptides thought to play a role in the pathological progression of Alzheimer disease (22). In normal biology, a variety of functions for APP have been implicated. The earliest to be described was the function of the secreted ectodomain of APP (sAPP) as protease nexin 2, a serine protease inhibitor that targets coagulation factor XIa (FXIa) with high specificity (23, 24). This inhibitory activity has been localized to the Kunitz domain of sAPP (25). Subsequently, sAPP has been found to stimulate growth in both neuronal and epithelial cells, to influence neurite outgrowth and synaptogenesis in the nervous system, and to stimulate motility in epithelial cells (26, 27). As an intact integral membrane protein, the structure of APP suggests similarity to membrane receptors (28), and recent data have suggested possible roles for APP as a cell surface receptor with signaling functions activated through cell-cell and cell-extracellular matrix contacts (26) and through proteolysis (29).
In the present study, we sought to identify specific substrate(s) of mesotrypsin. Hypothesizing that such substrates might possess canonical inhibitor domains, we employed an affinity-based proteomic screen designed to enrich for this class of putative mesotrypsin targets. We identify sAPP/protease nexin 2 as a protein that is highly expressed by prostate cancer cells and that is efficiently cleaved as a specific substrate by mesotrypsin. We further find that this cleavage has a large effect on the ability of sAPP/protease nexin 2 to act as an inhibitor of serine proteases, including FXIa.
EXPERIMENTAL PROCEDURES
Cell Culture
WPE1 NB11 and LNCaP cells were obtained from ATCC. Both are malignant, tumorigenic cell lines of human prostate epithelial origin. NB11 cells were cultured as described previously (30), in complete keratinocyte serum-free medium (Invitrogen) containing 50 μg/ml bovine pituitary extract (Invitrogen), 5 ng/ml epidermal growth factor supplement (Invitrogen), and 100 μg/ml gentamicin (Invitrogen). LNCaP cells were maintained in RPMI 1640 (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 100 μg/ml gentamicin until cells reached confluence, at which time the medium was replaced with serum-free RPMI 1640. Conditioned media for affinity purification, proteolysis experiments, and Western blotting were collected from cell cultures after 4–5 days, clarified by centrifugation, and stored on ice. Prior to SDS-PAGE and Western blotting, media were concentrated 5–7-fold using a Microcon concentrator (Millipore) with a 10,000 Mr cut-off.
Trypsin Affinity Chromatography of Conditioned Cell Culture Medium
The recombinant, catalytically inactive R117H,S195A mutant of human cationic trypsinogen was expressed, refolded, and purified as described previously (11). Purified R117H, S195A trypsinogen (30 mg) was proteolytically processed by 12 μg of bovine enteropeptidase (Roche Applied Science) in 80 ml of 100 mm HEPES, pH 8.0, 1 mm CaCl2 for 5 h at 37 °C. After confirming efficient processing by SDS-PAGE, the reaction was quenched with 1 mm phenylmethylsulfonyl fluoride for 1 h at 25 °C. Processed R117H,S195A trypsin was purified first on a HiTrap Benzamidine FF affinity column (GE Healthcare) by elution with a linear gradient from buffer A (50 mm HEPES, pH 8.0, 0.5 m NaCl) to buffer B (20 mm HCl). Subsequently, R117H,S195A trypsin was further purified to eliminate residual contamination with enteropeptidase on a HiLoad 16/60 Superdex 75 preparation grade gel filtration column (GE Healthcare) equilibrated and eluted with 50 mm HEPES, pH 8.0, 0.15 m NaCl, and 1 mm CaCl2. Following concentration by ultrafiltration using a Centriprep-10 concentrator (Amicon), purified R117H,S195A trypsin (8 mg) was adjusted to pH 6.5 with HCl and coupled to 1.2 ml of washed activated Affi-Gel-10 resin (Bio-Rad) overnight at 4 °C; unreacted groups were subsequently blocked by incubating for several h with dilute ethanolamine.
Proteins secreted by NB11 prostate epithelial cells and that possessed strong affinity to trypsin were enriched by chromatography of conditioned cell culture medium on the R117H, S195A trypsin affinity column. Medium was concentrated 8.75-fold using an Amicon stirred cell with a 10,000 Mr cut-off membrane and then dialyzed against 50 mm Tris, pH 7.6, and 0.2 m NaCl (Buffer A). The dialyzed protein was loaded onto the affinity column pre-equilibrated with buffer A, and then the column was eluted with a gradient to 20 mm HCl (buffer B) over 20 column volumes. Fractions containing proteins that eluted during the gradient were pooled and concentrated 20-fold using a Centriprep-10 concentrator (Millipore) prior to resolution by SDS-PAGE and protein identification by mass spectrometry.
Mass Spectrometry and N-terminal Sequencing
The mass spectrometry protein identification was performed essentially as previously described (31) at the Mayo Proteomics Research Center. Briefly, the 10% acrylamide SDS-polyacrylamide gel was silver-stained using the SilverSNAP stain for mass spectrometry kit (Pierce). Subsequently, the silver-stained gel bands were excised, destained, and then reduced and alkylated with dithiothreitol and iodoacetamide. Proteins were digested with trypsin (Promega) followed by peptide extraction with 60 μl of 2% trifluoroacetic acid, and then 60 μl of acetonitrile. Pooled extracts were concentrated and then brought up in 0.1% formic acid for protein identification by nanoflow liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analysis using a ThermoFinnigan LTQ Orbitrap hybrid mass spectrometer coupled to an Eksigent nanoLC-two-dimensional HPLC system. The MS/MS raw data were converted to DTA files by using ThermoFinnigan's Bioworks 3.1 and correlated to theoretical fragmentation patterns of tryptic peptide sequences from the Swiss Protein Database using both SEQUEST (ThermoElectron) and Mascot (Matrix Sciences) search algorithms running on a 10-node cluster.
Intact mass measurements were obtained using nanoflow liquid chromatography electrospray mass spectrometry (nanoLC-ESI-MS) at the Mayo Proteomics Research Center, again employing the ThermoFinnigan LTQ Orbitrap hybrid mass spectrometer coupled to the Eksigent nanoLC-two-dimensional HPLC system. The protein solutions were back-loaded onto a 250-nl OPTI-PAK trap (Optimize Technologies) custom packed with Michrom Magic C8 solid phase (Michrom Bioresources). Chromatography was performed using 0.2% formic acid in both the A solvent (98% water, 2% acetonitrile) and B solvent (80% acetonitrile, 10% isopropyl alcohol, 10% water); peaks were resolved with a 10% B to 50% B gradient over 20 min at 350 nl/min through a Michrom Magic C18 (75 μm × 150 mm) packed tip capillary column. The LTQ Orbitrap mass spectrometer was set to perform a Fourier transform full scan from 300 to 1800 m/z with resolution set at 60,000 (at 400 m/z). The Extract function in Excalibur was used to convert the raw mass spectra to monoisotopic masses. For protein samples that were reduced prior to nanoLC-ESI-MS, reduction involved adding 3 μl of 0.5 m tris(2-carboxyethyl)phosphine to 20 μl of the protein solution and heating at 55 °C for 20 min prior to injection. N-terminal sequencing of the HPLC-purified recombinant isolated Kunitz domain of APP (APPI) and its derivatives was performed on an automated protein sequencer (ABI-Procise cLC, Applied Biosystems) at the Mayo Proteomics Research Center.
Western Blotting
SDS-polyacrylamide gels (10% acrylamide) were electroblotted onto Immobilon-FL polyvinylidene difluoride membrane (Millipore) in a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad) according to the manufacturer's protocols. Membranes were blocked in 5% NFDM/TBST (nonfat dried milk/Tris-buffered saline with 0.5% Tween 20) overnight at 4 °C. For N-terminal sAPP detection, mouse monoclonal primary antibody 22C11 (Millipore catalog number MAB348) was used at a dilution of 1:1000. Membranes were incubated with primary antibody in blocking buffer for 2 h at 25 °C, washed three times in TBST, and then incubated at 25 °C for 1 h with a 1:80,000 dilution of peroxidase-conjugated goat anti-mouse secondary antibody (Jackson Immunoresearch Laboratories) in blocking buffer. Membranes were washed four times in TBST, developed using the ECL Plus Western blotting detection system (Amersham Biosciences) according to manufacturer protocols, and imaged on a ChemiDoc XRS molecular imager (Bio-Rad).
Production of Recombinant Human Proteases and APPI
Recombinant human cationic trypsinogen and mesotrypsinogen were expressed in Escherichia coli, isolated from inclusion bodies, refolded, purified, and activated with bovine enteropeptidase as described previously (11). Cationic trypsin and mesotrypsin concentrations were quantified by active-site titration using 4-nitrophenyl 4-guanidinobenzoate (Sigma) (32). Recombinant APPI was expressed in Pichia pastoris as a soluble secreted protein essentially as described previously (33). Expression cultures grown in BMMY medium (buffered medium containing methanol and yeast nitrogen base) at 30 °C were harvested after 48 h. The supernatant was subjected to 95% ammonium sulfate precipitation at room temperature. Protein pellets were resuspended in 20 mm Tris buffer, pH 7.8, and dialyzed overnight against 10 mm Tris buffer, pH 7.8, using 3500 Mr cut-off dialysis tubing (ThermoScientific). Dialyzed APPI samples were chromatographed on Q-Sepharose FF (GE Healthcare) and eluted with a gradient of 0–100% buffer B (50 mm Tris, pH 7.8, 1 m NaCl) and then further purified on a trypsin affinity column and eluted with a gradient of 0–100% buffer B (150 mm HCl). APPI concentrations were determined by titration with bovine trypsin (Sigma) as described previously (11).
Production, Purification, and Characterization of APPI* Cleaved at the Arg15-Ala16 Bond
Mesotrypsin-cleaved APPI* was produced on a preparative scale by digestion of 250 μm APPI with 5 μm mesotrypsin at 37 °C for 4 h, followed by acidification to pH 1 and purification from unreacted APPI by repeated chromatography on a 50 × 2.0-mm Jupiter 4μ 90-Å C12 column (Phenomenex) with a gradient of 30–85% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 0.6 ml/min. Purified APPI* was concentrated and acetonitrile was removed by dilution with distilled H2O and ultrafiltration in an Amicon Ultra-4 centrifugal filter unit with a 3000 Mr cut-off. APPI* concentrations were determined from integration of HPLC peak areas and comparison with standard curves generated from known quantities of titrated APPI.
Competitive Inhibition Studies
Concentrations of the chromogenic substrate benzyloxycarbonyl-Gly-Pro-Arg-p-nitroanalide (Z-GPR-pNA) (Sigma) were determined by an end point assay. For the study employing an octapeptide mimic of the APPI cleavage site, the peptide Gly-Pro-Ser-Arg-Ala-Met-Ile-Tyr, acetylated at the N terminus and featuring a C-terminal amide to better mimic an internal peptide sequence, was synthesized by standard solid-phase methods and HPLC-purified to 98.6% purity by EZBiolabs. The lyophilized peptide was dissolved in 10 mm NaOAc, pH 6.0, prior to use. Peptide concentration was determined in 0.1 n NaOH by absorbance at 293 nm using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), using a calculated extinction coefficient of 2400 m−1 cm−1 according to the supplier's recommendations. Working stocks of enzyme, substrate, and inhibitors were prepared for competitive inhibition studies as described previously (11). Assays were conducted at 37 °C in a Varian Cary-100 spectrophotometer equipped with a multicell transporter and a circulating water bath. Assay buffer (740 μl; 100 mm Tris-HCl, pH 8.0, 1 mm CaCl2), substrate (20 μl), and inhibitor (20 μl) were mixed and equilibrated in cuvettes prior to reaction initiation by the addition of enzyme (20 μl). Reactions were followed spectroscopically for 3–5 min, and initial rates were determined from the absorbance increase caused by the release of p-nitroaniline (ϵ410 = 8480 m−1 cm−1) (34). Data were globally fitted by multiple regression to Equation 1, the classic competitive inhibition equation, using Prism (GraphPad Software). Reported inhibition constants are average values obtained from multiple independent experiments.
 |
Slow, Tight Binding Inhibition Studies
Slow, tight binding inhibition of cationic trypsin by APPI was measured in a manner similar to that described previously for bovine pancreatic trypsin inhibitor (BPTI) inhibition (11). Briefly, reactions were initiated by the addition of enzyme (0.1 nm) to quartz cuvettes containing substrate Z-GPR-pNA (150 μm) and a range of APPI concentrations (10–50 nm). Reactions were run at 25 °C and were followed spectroscopically for 12 h so that reliable steady-state rates could be obtained. Conversion of substrate to product did not exceed 10% over the reaction time course. Data were analyzed as described previously (11, 35) using Equation 2,
where vi and v0 are the steady-state rates in the presence and absence of inhibitor, Km is the Michaelis constant for cationic trypsin cleavage of Z-GPR-pNA, and [S0] and [I0] are the initial concentrations of substrate and inhibitor. Equation 2 predicts that a plot of (v0 − vi)/vi versus [I0] will yield a straight line passing through the origin, with a slope of 1/Ki (1 + [S0]/Km), allowing the calculation of Ki (35, 36). For these calculations, we used a Km value of 36.5 μm, as determined previously (11). The reported Ki value represents the average of six independent experiments.
APPI Hydrolysis Studies
The depletion of intact APPI in time course incubations with active mesotrypsin or cationic trypsin was monitored by HPLC and/or 16% SDS-Tricine polyacrylamide gel (37, 38). Enzyme incubations with APPI were carried out in 0.1 m Tris-HCl, pH 8.0, and 1 mm CaCl2 at 37 °C. Aliquots for HPLC analysis were withdrawn at periodic intervals, quenched by acidification to pH ≤1, and then frozen at −20 °C until analyzed. Mesotrypsin or cationic trypsin, APPI, and hydrolysis products were resolved on a 50 × 2.0-mm Jupiter 4μ 90 Å C12 column (Phenomenex) with a gradient of 0–100% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 0.6 ml/min over 50 min. Peak integration to quantify the disappearance of intact APPI over time was carried out as described previously for BPTI (11). Initial rates were obtained by linear regression using a minimum of six data points within the initial linear phase of the reaction; the hydrolysis rates reported represent the average of three independent experiments.
Factor XIa Inhibition and Activated Partial Thromboplastin Time (APTT) Assays
To obtain apparent Ki values for inhibition of FXIa (Hematologic Technologies, Inc.) by APPI or cleaved APPI*, FXIa was incubated with increasing concentrations of inhibitor for 30 min at 37 °C. Residual enzyme activity was measured under pseudo-first order kinetic conditions as previously reported (25, 33), using the substrate t-butoxycarbonyl-Glu(γ-benzyl ester)-Ala-Arg-4-methylcoumaryl-7-amide (Peptides International), and the concentration at which 50% of activity remains (IC50) was determined using Kaleidograph version 3.05 (Abelbeck Software, PCS, Inc., Reading, PA) non-linear least squares regression software. The IC50 value was converted to an inhibition constant (Ki) using Equation 3,
where [S] represents the substrate concentration, and Km is the Michaelis constant of FXIa for t-butoxycarbonyl-Glu(γ-benzyl ester)-Ala-Arg-4-methylcoumaryl-7-amide, determined to be ∼275 μm. To measure the effect of APPI or cleaved APPI* on clotting time, APTT assays were performed as described previously, using 25 μl of APTT reagent (Kontact, Fisher) (33).
RESULTS
Identification of APP as a Mesotrypsin Substrate
Relative to other trypsins, mesotrypsin displays reduced activity toward classic protein substrates of trypsin, including pancreatic zymogens (7) and protease-activated receptors (8–10), yet displays enhanced catalytic activity toward one category of protein substrates, the canonical protease inhibitors (7, 11). We hypothesized that as yet unidentified physiological substrates of mesotrypsin are likely to be canonical serine protease inhibitors capable of binding very tightly to trypsin, and we designed a screen to identify such mesotrypsin substrates. Because we have found transcription of the PRSS3 gene encoding mesotrypsin to be up-regulated in prostate cancer cells,2 we screened for novel mesotrypsin substrates produced by prostate cancer cells. First, we enriched conditioned medium from WPE1 NB11 prostate cells for trypsin inhibitors by using trypsin affinity chromatography. Many canonical inhibitor domains are components within large multidomain proteins; to minimize proteolytic fragmentation of such proteins, we employed an affinity resin made with a mutant of human cationic trypsin that is catalytically inactive yet retains normal affinity for canonical trypsin inhibitors. Using SDS-PAGE, we compared samples of enriched conditioned medium that were either untreated or treated with mesotrypsin (Fig. 1A). We identified several prominent protein bands that were present before treatment but absent after treatment with mesotrypsin. These bands were excised from a silver-stained gel, digested with trypsin, and subjected to mass spectrometry for protein identification. One of the most intensely stained bands in the untreated sample, with an apparent molecular weight of ∼120,000, was identified as APP, with 33 unique peptides giving 48% sequence coverage of the complete APP coding sequence (Fig. 1B). The farthest N-terminal (LEVPT…) and C-terminal (…VHHQK) sequence matches correspond to the N and C termini, respectively, of soluble APP-α (sAPPα), secreted through the activity of α-secretase. This protein, also known as protease nexin 2, contains a Kunitz serine protease inhibitor domain (sequence shown in red in Fig. 1B), which has been found to strongly inhibit trypsin as well as several enzymes of the coagulation system (23, 24). In the mesotrypsin-treated sample, the 120 kDa band is depleted, whereas two newly appearing bands with apparent molecular mass of ∼40 and ∼65 kDa were found by trypsin digestion and nanoLC-MS/MS to represent cleavage fragments of APP. The 40 kDa band yielded primarily peptides mapping to the N-terminal half of APP, whereas the 65 kDa band yielded predominantly C-terminally mapping peptides (not shown). The sizes of these fragments are roughly consistent with cleavage at the reactive site Arg within the Kunitz domain (capitalized in Fig. 1B).
FIGURE 1.
Identification of APP as a mesotrypsin substrate. A, conditioned medium from NB11 prostate cancer cells was enriched for trypsin-binding proteins by affinity chromatography as described under “Experimental Procedures.” Resultant protein samples, either untreated (left lane) or subjected to digestion with 50 nm mesotrypsin at 37 °C for 1 h (right lane), were resolved by SDS-PAGE and silver-stained, and bands differing between the two lanes were identified by nanoLC-MS/MS as described under “Experimental Procedures.” The band labeled APP was identified with 33 unique peptide matches to the human APP sequence; bands labeled B40 and B65 were tentatively identified as N-terminal and C-terminal proteolytic fragments derived from APP. B, the complete sequence of the human APP gene, showing coverage of peptides identified by nanoLC-MS/MS in boldface type and underlined. The Kunitz domain sequence is shown in red, with the reactive site Arg residue capitalized.
To determine whether mesotrypsin would target sAPP/protease nexin 2 for selective cleavage in the context of a more complex biological sample, we assessed samples of mesotrypsin-treated crude concentrated conditioned media from different cell lines by Western blotting (Fig. 2). Consistent with the sizes of cleavage fragments identified above, we found that an antibody recognizing the N terminus of APP detected the full-length protein at ∼120 kDa and a fragment of ∼40 kDa. In incubations with increasing amounts of mesotrypsin, we observed a concentration-dependent depletion of intact sAPP/protease nexin 2 and a corresponding increase in signal intensity for the cleavage fragment; substantial cleavage was observed at mesotrypsin concentrations as low as 1 nm (Fig. 2, A and B). Control experiments confirmed that APP cleavage was due to mesotrypsin proteolytic activity because no cleavage was detected in incubations with mesotrypsin inactivated by the small molecule covalent inhibitor phenylmethylsulfonyl fluoride or with a catalytically inactive mesotrypsin-S195A mutant (Fig. 2C). Furthermore, mesotrypsin displayed unique selectivity in targeting APP for cleavage because the highly homologous human cationic trypsin at similar concentration showed no evidence of cleaving APP (Fig. 2C).
FIGURE 2.
Hydrolysis of sAPP by mesotrypsin. Conditioned media from prostate cancer cell lines were treated with mesotrypsin and analyzed for APP protein on Western blots. Serum-free conditioned media from confluent WPE1-NB11 cells (A) or LNCaP cells (B) were collected after 5 days, concentrated 6-fold, and treated for 1 h at 37 °C with recombinant mesotrypsin at the concentrations shown. Western blots probed with antibody 22C11, which recognizes an N-terminal APP epitope, show a mesotrypsin-dependent disappearance of intact sAPP (∼120 kDa), along with concomitant accumulation of a cleavage fragment of ∼40 kDa. C, incubation of WPE1-NB11 conditioned medium was performed as described in A with the following recombinant enzymes: buffer only (lane 1), mesotrypsin inhibited with 10 mm phenylmethylsulfonyl fluoride (lane 2), mesotrypsin-S195A (lane 3), cationic trypsin (lane 4), and mesotrypsin (lane 5). All enzymes were at 100 nm concentration.
Characterization of the Cleavage Site Using the Recombinant Kunitz Domain of APP
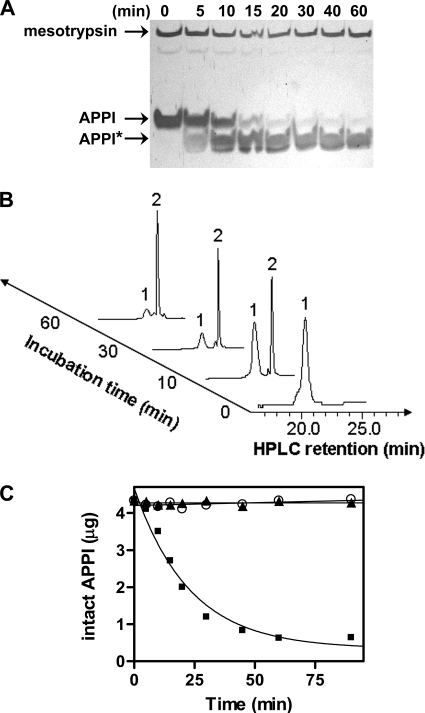
Suspecting that the site of sAPP/protease nexin 2 cleavage by mesotrypsin would lie within the Kunitz protease inhibitor domain, we pursued studies using APPI. In time course experiments following the proteolysis of APPI by SDS-PAGE (Fig. 3A) or by HPLC analyses (Fig. 3B), we found that mesotrypsin cleaved APPI to produce a single stable species. With extended incubation times, it appeared that the cleavage reaction did not proceed to completion but formed an equilibrium in which a small population of residual intact APPI remained present (Fig. 3C). This observation is consistent with previous studies of other canonical serine protease inhibitors, wherein the inhibitors have been observed to bind very tightly to cognate proteases in a substrate-like manner, positioning a specific peptide bond (the “reactive site”) in the active site of the protease (12, 13). An inhibited enzyme may eventually cleave the bound inhibitor, although at a rate many orders of magnitude slower than hydrolysis of a normal substrate, to produce a “modified” inhibitor species singly cleaved at the reactive site but held together by disulfide bonds; by convention, this species is indicated with an asterisk. The modified inhibitor may also bind to the enzyme and may undergo enzymatic religation, resulting in an equilibrium mixture of intact and modified species (12, 13). Given the data shown in Fig. 3, we hypothesized that mesotrypsin interacts with APPI in a manner similar to that of an effectively inhibited protease but with weaker affinity and with an accelerated rate of cleavage of the reactive site peptide bond, resulting in rapid formation of an APPI*/APPI equilibrium. Under our reaction conditions, the APPI*/APPI equilibrium stabilized at a ratio of ∼9:1 (Fig. 3C). Proteolysis of APPI was uniquely catalyzed by active mesotrypsin because no cleavage was observed upon incubation with the same concentration of either the mesotrypsin-S195A mutant or active cationic trypsin (Fig. 3C).
FIGURE 3.
Hydrolysis of recombinant APPI by mesotrypsin. A, SDS-polyacrylamide gel shows a time course of APPI hydrolysis by mesotrypsin. APPI (25 μm) and mesotrypsin (250 nm) were incubated at 37 °C, and then samples were withdrawn and quenched at various time points for subsequent analysis. The arrows indicate mesotrypsin (24 kDa), APPI (6.3 kDa), and APPI* hydrolysis products. B, representative HPLC chromatograms are shown from a similar time course of APPI hydrolysis by mesotrypsin. Peak 1 eluting at 21.5 min and peak 2 eluting at 23.0 min were isolated and subjected to mass spectrometry and N-terminal sequencing to confirm that they represent intact APPI and cleaved APPI*, respectively (see Fig. 4 and Table 1). C, disappearance of intact APPI was quantified by integration of HPLC peak 1 in a time course similar to that illustrated in B, in incubations of APPI with mesotrypsin (■), the catalytically inactive mesotrypsin-S195A (○), or cationic trypsin (▴). Equilibrium was attained between intact and cleaved APPI over the course of the 90-min incubation with mesotrypsin. APPI concentration was 25 μm, and enzyme concentration was 250 nm in each incubation.
To identify the proteolysis products resolved by HPLC in Fig. 3B, we isolated peaks 1 and 2 and obtained intact mass measurements and N-terminal sequence of these species both before and after reduction; the results of these studies are reported in Table 1. As shown in Fig. 4, intact APPI is a 57-amino acid peptide featuring three internal disulfide bonds and the N-terminal sequence EVCSE. The reactive site bond identified in crystal structures of APPI bound to FXIa (33) and trypsin (39, 40) is located between Arg13 and Ala14 (indicated by a black arrow in Fig. 4); from here on, we will refer to this bond as the Arg15-Ala16 bond, according to its alignment with the archetypal Kunitz family inhibitor BPTI and for consistency with the bulk of prior literature. Cleavage at the reactive site Arg15-Ala16 bond creates a new species (APPI*) composed of two peptide chains joined by two disulfide bonds; because hydrolysis results in incorporation of a water molecule, the intact mass of APPI* is 18 Da heavier than that of APPI. Using nanoLC-ESI-MS, we obtained intact mass measurements within 15 ppm of the predicted theoretical masses for nonreduced and reduced APPI, nonreduced APPI*, and the anticipated C-terminal cleavage fragment isolated from APPI* (Table 1). Furthermore, for APPI*, we obtained overlapping N-terminal sequence reflective of the dual amino termini generated by cleavage at the Arg15-Ala16 bond, whereas the reduced and isolated C-terminal cleavage fragment yielded the N-terminal sequence AMISR, again fully consistent with the predicted Arg15-Ala16 cleavage site (Table 1). Thus, our results clearly establish that the site at which mesotrypsin cleaves APPI is the same reactive site that binds to and inhibits other serine proteases.
TABLE 1.
Intact mass and N-terminal sequence results from APPI and products
| NanoLC-ESI-MS observed mass | Theoretical mass | N-terminal sequence(s) | |
|---|---|---|---|
| Da | Da | ||
| APPI (peak 1) | 6262.68116 | 6262.59158 | EVCSE … |
| APPI (peak 1), reduced | 6268.72923 | 6268.63853 | |
| Mesotrypsin-cleaved APPI* (peak 2) | 6280.69339 | 6280.60214 | EVSCE … /AMISR … |
| C-terminal fragment resolved from reduced APPI* | 4879.13654 | 4879.07323 | AMISR … |
FIGURE 4.
Scheme for identification of Arg15-Ala16 as the mesotrypsin cleavage site within APPI. The complete sequence of APPI is shown, with intramolecular disulfide bonds indicated by black brackets below the sequence. The Arg15-Ala16 reactive site bond, anticipated to be specifically targeted for cleavage by mesotrypsin, is indicated by the black arrow. As illustrated by the scheme, mesotrypsin cleavage yields a single species composed of two peptide chains covalently linked by two disulfide bonds; because a water molecule has been incorporated in the enzymatic hydrolysis reaction, this APPI* species is 18 Da higher in mass than the APPI precursor. Following reduction of the disulfides, the peptide chains can be isolated by HPLC and individually characterized. The depicted species APPI (HPLC peak 1 from Fig. 3B), APPI* (HPLC peak 2 from Fig. 3B), and the C-terminal fragment were isolated and analyzed for intact mass and N-terminal sequence, with the results shown in Table 1.
Characterization of Cleavage Kinetics Using the Recombinant Kunitz Domain of APP
Next, we sought to determine the kinetics of cleavage, which would describe whether the mesotrypsin/APPI interaction is more substrate-like or inhibitor-like. Testing APPI as an inhibitor of mesotrypsin versus the small chromogenic peptide substrate Z-GPR-pNA, we observed a classic competitive pattern of inhibition, with an inhibition constant Ki of 136 ± 19 nm (Fig. 5A). The competitive inhibition equation used to determine Ki also describes the enzymatic reaction of one substrate in the presence of an alternative, competing substrate, with the added condition that the observed Ki for the alternative substrate must be equivalent to its Km (41). Thus, the Km for the cleavage of APPI at Arg15-Ala16 by mesotrypsin is 136 nm.
FIGURE 5.
Kinetics of APPI inhibition of and hydrolysis by mesotrypsin. A, mesotrypsin cleavage of peptide substrate Z-GPR-pNA is competitively inhibited by APPI with a Ki of 136 nm; because APPI is an alternative, competing substrate for mesotrypsin, this value also represents the Km for mesotrypsin cleavage of APPI. APPI concentration was 0 nm (□), 50 nm (▾), 100 nm (○), 200 nm (■), or 400 nm (▵); mesotrypsin concentration was 0.25 nm. Data were fit globally to the competitive inhibition equation; the Lineweaver-Burk double reciprocal transform is shown here. B, mesotrypsin cleavage of Z-GPR-pNA is only minimally inhibited by 1 mm Gly-Pro-Ser-Arg-Ala-Met-Ile-Tyr, a peptide mimic of the APPI binding loop. Peptide concentration was 0 mm (▵) or 1 mm (■); mesotrypsin concentration was 0.25 nm. C, disappearance of intact APPI in a time course similar to that illustrated in Fig. 3B was quantified by HPLC peak integration. The linear initial rate of hydrolysis, observed under conditions of enzyme saturation, allows calculation of a catalytic rate constant kcat of 4.2 × 10−2 s−1. APPI and mesotrypsin concentrations were 25 μm and 25 nm, respectively. D, disappearance of intact APPI in a time course incubation with cationic trypsin was quantified by HPLC peak integration. The linear initial rate of hydrolysis, observed under conditions of enzyme saturation, allows calculation of a catalytic rate constant kcat of 1.8 × 10−5 s−1. APPI and cationic trypsin concentrations were 50 μm and 5 μm, respectively.
We based our screen to identify mesotrypsin substrates on the hypothesis that the specificity of mesotrypsin derives in part from a preference for proteolysis sites stabilized in a canonical inhibitor-like conformation. This hypothesis predicts that presentation of the cleavage site of APP in the context of the Kunitz domain tertiary structure may be an important factor in promoting recognition and cleavage by mesotrypsin. To experimentally determine whether this is the case, we tested the interaction of mesotrypsin with an octapeptide designed to mimic the stretch of primary sequence that contains the reactive site of APPI. Crystal structures indicate that the majority of contacts between APPI and trypsins are formed by the P4–P3′ residues Gly-Pro-Cys-Arg-Ala-Met-Ile (39, 40). We synthesized a peptide recapitulating this sequence, but substituting the P2 Cys with Ser to prevent peptide dimerization and adding a Tyr residue at the P4′ C terminus to facilitate peptide quantification. We measured the ability of this peptide to act as an inhibitor of mesotrypsin, using Z-GPR-pNA as a substrate. At submillimolar concentrations, we observed no inhibition of mesotrypsin activity (not shown), whereas at 1 mm peptide, we observed only a very slight degree of inhibition (Fig. 5B), suggesting that the inhibition constant Ki for the peptide must be substantially higher than 1 mm. Comparing the affinity of mesotrypsin toward the peptide with that toward APPI, it appears that presentation of the reactive site sequence within the context of the Kunitz domain converts a target with very poor affinity into a target with high nanomolar affinity; this observation is consistent with the idea that mesotrypsin may preferentially target protein substrates constrained in a canonical conformation.
We next used a direct HPLC assay to assess the rate of APPI proteolysis by mesotrypsin. By measuring APPI hydrolysis by mesotrypsin under saturating conditions where the APPI concentration vastly exceeded Km, we determined a linear initial rate of hydrolysis corresponding to a catalytic rate constant of 0.042 ± 0.003 s−1 (Fig. 5C). This rate constant corresponds to a catalytic turnover time of 24 s, indicating that APP hydrolysis by mesotrypsin could represent a significant physiological process on the biological time scale. This represents a large rate acceleration compared with cleavage of canonical inhibitors by most serine proteases, which typically proceed with turnover times measured in days or months (e.g. see Refs. 11 and 35). For a directly relevant comparison, we used similar methods to measure the rate of APPI hydrolysis by cationic trypsin; this experiment required a 200-fold higher enzyme concentration and a much longer time course for detecting cleavage. The linear initial rate of APPI hydrolysis by cationic trypsin corresponded to a catalytic rate constant of 1.8 ± 0.1 × 10−5 s−1, equivalent to a turnover time of ∼15 h (Fig. 5D). Thus, the catalytic rate constant of APPI cleavage by mesotrypsin is accelerated more than 2,300-fold by comparison with cationic trypsin.
Mesotrypsin Cleavage Severely Compromises the Function of the APP Kunitz Domain as an Inhibitor
We next hypothesized that cleavage by mesotrypsin might play a role in regulating the serine protease inhibitory activity of APP/protease nexin 2. To assess the effect of cleavage on inhibitory activity, we purified mesotrypsin-cleaved APPI* by HPLC and compared the ability of intact and cleaved APPI to inhibit human cationic trypsin. Initially, we tested APPI inhibition of cationic trypsin using the classic competitive inhibition model (not shown). These experiments revealed that the inhibition constant Ki was less than 1 nm, which in our study design reflected the lower limit of resolution for which this treatment would be valid. To enable accurate measurement of Ki using a slow, tight binding model of inhibition, we turned to an alternative study design described under “Experimental Procedures,” which yielded an inhibition constant Ki of 1.7 ± 0.6 × 10−10 m (Fig. 6, A and B). This value is very close to previously reported Ki values for isolated APP Kunitz domains toward bovine and porcine trypsins (42, 43). For purified cleaved APPI*, by contrast, the standard competitive inhibition model proved to be an adequate treatment, yielding an inhibition constant Ki of 2.2 ± 0.3 × 10−8 m (Fig. 6C). Thus, mesotrypsin cleavage of APPI at the reactive site reduces its affinity for cationic trypsin by 2 orders of magnitude.
FIGURE 6.
APPI* cleaved at Arg15-Ala16 is severely compromised as an inhibitor of cationic trypsin. A and B, intact APPI inhibited cationic trypsin as a slow, tight binding inhibitor with a Ki of 170 pm. A, steady-state equilibrium rates were obtained from reactions including various concentrations of APPI as noted on the plot; reactions included 0.1 nm cationic trypsin and 150 μm Z-GPR-pNA substrate. B, The replot of (v0 − vi)/vi versus [APPI], where v0 is the uninhibited rate and vi is the rate in the presence of APPI, allows calculation of Ki as described under “Experimental Procedures.” C, cleaved APPI* is a classic competitive inhibitor of cationic trypsin, with an inhibition constant Ki of 22 nm. APPI* concentration was 0 nm (□), 10 nm (▾), 25 nm (○), or 50 nm (■); cationic trypsin concentration was 0.8 nm. Data were fitted as in Fig. 5A.
Because FXIa is a particularly important physiological target regulated by sAPP/protease nexin 2 in vivo (23–25), we next sought to determine the effect of mesotrypsin cleavage on the ability of APPI to inhibit FXIa. Residual FXIa activity was measured following pre-equilibration with increasing concentrations of either intact APPI or cleaved APPI*, allowing determination of equilibrium binding constants Ki of 6.3 ± 1.3 × 10−10 m for intact APPI (Fig. 7A) and 6.6 ± 1.1 × 10−8 m for cleaved APPI* (Fig. 7B). Thus, similar to our finding for cationic trypsin, the inhibition of FXIa by APPI is weakened by 2 orders of magnitude upon cleavage of APPI by mesotrypsin. In parallel with in vitro kinetic assays, we measured the effects of APPI and APPI* inhibition in a biological assay of FXIa function, the APTT assay, which monitors the activity of the intrinsic pathway of blood coagulation. In this study, we found that cleaved APPI* was strikingly less effective at prolonging clotting time as compared with intact APPI tested over the same concentration range (Fig. 7C).
FIGURE 7.
APPI* cleaved at Arg15-Ala16 is severely compromised as an inhibitor of factor XIa. Equilibrium binding constants were determined by preincubating FXIa (25 pm) with increasing concentrations of intact APPI (A) or cleaved APPI* (B) for 30 min at 37 °C and then measuring the remaining FXIa activity toward t-butoxycarbonyl-Glu(benzyl ester)-Ala-Arg-4-methylcoumaryl-7-amide (275 μm). Points represent mean values ± S.D. of four assays each. C, effects of APPI and APPI* on the clotting time in an APTT assay are compared. Values are reported as -fold increase compared with controls in the absence of inhibitors; points represent mean values ± S.D. of four determinations for APPI (solid circles) and five determinations for APPI* (solid squares). Assays were carried out in microcuvettes at 37 °C using normal pooled plasma.
DISCUSSION
Mesotrypsin is catalytically impaired for cleavage of most protein substrates by comparison with cationic and anionic trypsins (7). A prominent exception, for which mesotrypsin appears to possess enhanced catalytic capability compared with its proteolytic siblings, is in the cleavage of canonical protease inhibitors (7, 11). Here, by pursuing the hypothesis that the natural substrates of mesotrypsin may be endogenous canonical trypsin inhibitors, we have identified APP/protease nexin 2 as a protein that is proteolyzed in a highly efficient manner by mesotrypsin. We have identified the mesotrypsin cleavage site as the Arg15-Ala16 reactive site bond of the APP Kunitz protease inhibitor domain, and we have also shown that this cleavage drastically diminishes the effectiveness of the APP Kunitz domain as an inhibitor of serine proteases.
To evaluate the likelihood that APP/protease nexin 2 cleavage may be a significant activity of mesotrypsin in vivo, we have defined the steady-state kinetic parameters governing this proteolytic event. The specificity constant kcat/Km for the reaction lies well within the range observed for other chymotrypsin family serine proteases toward specific physiological protein substrates (Table 2). This, in itself, is not unusual for a canonical inhibitor/target protease inhibitory interaction; these inhibitors are frequently hydrolyzed with kcat/Km values that are quite high (104 to 106 m−1 s−1) but with kcat and Km values that individually are many orders of magnitude lower than those for normal substrates, reflecting an interaction in which inhibitors bind to proteases extremely tightly but are cleaved extremely slowly (16). Mesotrypsin binds to APPI with a Km of 140 nm, which is quite low for a protease-substrate interaction but not unprecedented among highly selective proteases, as in the case of tissue-type plasminogen activator (t-PA), which binds to its primary physiological target plasminogen in the presence of fibrin with a Km of 18 nm (44). Mesotrypsin cleaves APPI with a kcat of 0.042 s−1, corresponding to an enzymatic turnover time of 24 s. Although quite slow, this value is only 5-fold slower than that for the physiologically relevant cleavage of plasminogen by t-PA (44). We conclude that the steady-state kinetic parameters for cleavage of APPI by mesotrypsin are consistent with a possible physiological role for this reaction. Like t-PA (45) and in contrast with less specific proteases like cationic trypsin, mesotrypsin may have evolved a reduced rate of catalysis along with greater stringency in the determinants of productive binding to achieve selective proteolysis of a limited number of substrates. The present study suggests that APP/protease nexin 2 is likely to be such a specific physiological substrate of mesotrypsin.
TABLE 2.
Kinetic constants for selected chymotrypsin family serine proteases toward specific protein substrates
| Enzyme | Substrate | Km | kcat | kcat/Km | Source/Reference |
|---|---|---|---|---|---|
| m | s−1 | m−1s−1 | |||
| Mesotrypsin | APPI | 1.4 × 10−7 | 0.042 | 3.0 × 105 | Present study |
| Cationic trypsin | Chymotrypsinogen A | 2.1 × 10−5 | 2.3 | 1.1 × 105 | Ref. 46 |
| Enteropeptidase | Cationic trypsinogen | 1.4 × 10−6 | 35 | 2.5 × 107 | Ref. 47 |
| t-PA | Lys-plasminogen | 7.6 × 10−6 | 0.2 | 2.9 × 104 | Ref. 44 |
| t-PA | Lys-plasminogen + fibrin | 1.8 × 10−8 | 0.2 | 1.2 × 107 | Ref. 44 |
| u-PAa | Lys-plasminogen | 5.4 × 10−6 | 1.7 | 3.1 × 105 | Ref. 48 |
a Urokinase-type plasminogen activator.
We found that the three-dimensional structure of APPI is critical for mesotrypsin recognition and binding, since an unstructured peptide mimic of the APPI binding loop associated with mesotrypsin >10,000-fold more weakly (Fig. 5, A and B). In a previous structural study of the mesotrypsin complex with the canonical inhibitor BPTI, we found that mesotrypsin Arg-193, a residue not possessed by other trypsins and important to the inhibitor resistance exhibited by mesotrypsin (7, 49), undergoes conformational changes to accommodate inhibitor binding, packing tightly into a crevice on the enzyme surface and becoming much more highly ordered upon complex formation (11). We hypothesize that this conformational change may be a general requirement for mesotrypsin to bind a protein substrate and that protein sequences preconfigured in the canonical conformation might be especially effective at inducing the change, resulting in a “conformational specificity” of mesotrypsin toward substrates possessing an appropriate cleavage site stabilized in the conformation typified by the canonical inhibitor binding loop. The concept of conformational specificity, that the most rapidly hydrolyzed substrates of a protease would be those with relatively rigid local tertiary structure in the optimum conformation for binding to the enzyme, was at one time proposed as a general feature of the chymotrypsin family proteases (50). Subsequently accumulated structural data have shown this not to be the general case, because many proteases seem to prefer flexible cleavage sites that undergo considerable conformational reorganization upon protease-substrate complex formation (51), and engineering of a protein substrate cleavage site for enhanced flexibility has been seen to improve protease efficiency (52). Mesotrypsin appears to be an exception to the general rule, a protease for which the unstructured substrates preferred by other enzymes may bind too weakly for efficient cleavage, whereas canonical inhibitors, which bind other proteases too tightly for efficient cleavage and release, become the ideal specific substrates for mesotrypsin. The use of conformation and protein context as a determinant of substrate specificity is again reminiscent of the highly selective protease t-PA, which binds plasminogen many orders of magnitude more tightly than peptide mimics (44), due at least in part to steric restrictions of the enzyme active site that favor local conformational properties of the natural protein substrate (53).
Having identified sAPP/protease nexin 2 as a mesotrypsin substrate, it is pertinent to consider the potential repercussions of this proteolytic event in vivo. As a protease inhibitor, the primary target of sAPP/protease nexin 2 is believed to be FXIa, a coagulation factor activated by thrombin on the platelet surface, which then serves an essential function in activation of factor IX and the propagation of the coagulation response, leading to increased production of active thrombin (54). Release of high concentrations of sAPP/protease nexin 2 by activated platelets is probably an important physiological mechanism for regulation of the procoagulant/anticoagulant balance in the vicinity of a developing clot (25). Cancer progression is abetted by a hypercoagulable state, in which tumor cell-produced tissue factor initiates the coagulation cascade, leading to thrombin activation, thrombin signaling through protease activated receptors, platelet-tumor aggregation, tumor adhesion to endothelium and subendothelial matrix, metastasis, tumor growth, and tumor angiogenesis (55, 56). Although sAPP/protease nexin 2, either tumor-produced (57–60) or derived from platelets or plasma, would be expected to counteract this procoagulant state, tumor cell-produced mesotrypsin, by disabling the sAPPα/protease nexin 2 Kunitz domain, may contribute to the prothrombotic nature of the tumor microenvironment and enhance malignant progression. Relevant to this possibility, overexpression of the PRSS3 gene encoding mesotrypsin has been reported to significantly enhance transendothelial migration of non-small cell lung carcinoma cells (5). In another intriguing report, a highly metastatic subclone of the pancreatic carcinoma cell line SUIT-2 was found to secrete a ∼43-kDa protein detected by the same N-terminal APP antibody employed in our study (58); this may represent a product of APP cleavage by endogenously produced mesotrypsin. In the present study, we have shown that cleavage by mesotrypsin substantially compromises one of the functions of APP, the protease inhibitory activity of sAPP/protease nexin 2, an activity important in the regulation of the coagulation pathway. It may be that by disabling this activity, mesotrypsin also shifts the balance of sAPP function toward its alternative activities as a growth factor and motogenic factor (27), in essence acting as a regulatory switch. Cleavage of sAPP by mesotrypsin might also influence its stability and longevity, because the reuptake and cellular catabolism of sAPP are mediated through binding to the low density lipoprotein receptor-related protein in a high affinity interaction that is dependent upon the Kunitz inhibitor domain (61). Evaluation of these possibilities and definition of the full impact of mesotrypsin on the integrated functions of APP will be an important direction for future research.
Finally, although the present study used epithelial prostate cancer cells to investigate substrates of mesotrypsin, our identification of APP as a mesotrypsin substrate may also be relevant to the brain. Trypsinogen 4, a splice isoform of mesotrypsinogen incorporating an alternative exon 1, is highly expressed in the brain (62–64). Proteolytic activation of trypsinogen 4 produces an active enzyme identical in protein sequence and catalytic properties to pancreatic mesotrypsin; in the context of the brain, this enzyme has also been referred to as trypsin 4, trypsin IV, or brain trypsin (62–64). The inhibitory activity of sAPP/protease nexin 2 plays a key role in regulating cerebral hemostasis and preventing thrombosis during cerebral vascular injury (65–67). Thus, in the brain as elsewhere, by attacking and cleaving the inhibitory Kunitz domain of sAPP/protease nexin 2, mesotrypsin may modulate the procoagulant/anticoagulant balance. Cleavage of APP by mesotrypsin may also have functional implications for other processes of the nervous system in which the APP Kunitz domain has been implicated. For example, neurite outgrowth is stimulated by cell-cell adhesion involving cell surface-expressed APP; cellular substrata displaying APP isoforms that possess the Kunitz domain are most effective at promoting neurite outgrowth (68). Furthermore, structural models of intact APP770 place the Kunitz domain in a fully exposed position on the side of the ectodomain farthest from the membrane attachment point (69). Thus, it is plausible that intermolecular interactions involving the Kunitz domain may impact some of the adhesion properties of cell surface APP and that cleavage of the Kunitz domain by mesotrypsin could impact these properties as well. The identification of the APP Kunitz domain as a highly specific substrate of mesotrypsin calls for further studies that will elucidate the functional consequences of this cleavage event both in epithelial tissues and tumors and in the nervous system.
This work was supported, in whole or in part, by National Institutes of Health Grants P50 CA091956-08 (to E. S. R.) and HL74124 and HL46213 (to P. N. W.). This work was also supported by Bankhead-Coley Florida Biomedical Research Program Award 07BN-07 (to E. S. R.).
A. Hockla, D. Radisky, and E. S. Radisky, manuscript in preparation.
- APP
- amyloid precursor protein
- APPI
- recombinant Kunitz inhibitor domain of APP
- FXIa
- factor XIa
- sAPP
- secreted ectodomain of APP
- APPI*
- two-chain APPI cleaved at the Arg15-Ala16 bond
- Z
- benzyloxycarbonyl
- pNA
- p-nitroanalide
- nanoLC
- nanoflow liquid chromatography
- MS/MS
- tandem mass spectrometry
- ESI-MS
- electrospray ionization mass spectrometry
- HPLC
- high pressure liquid chromatography
- APTT
- activated partial thromboplastin time
- BPTI
- bovine pancreatic trypsin inhibitor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- t-PA
- tissue-type plasminogen activator.
REFERENCES
- 1.Chen J. M., Ferec C. (2000) Pancreas 21, 57–62 [DOI] [PubMed] [Google Scholar]
- 2.Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S. M., Aburatani H. (2005) Genomics 86, 127–141 [DOI] [PubMed] [Google Scholar]
- 3.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Lancet D., Shmueli O. (2005) Bioinformatics 21, 650–659 [DOI] [PubMed] [Google Scholar]
- 5.Diederichs S., Bulk E., Steffen B., Ji P., Tickenbrock L., Lang K., Zänker K. S., Metzger R., Schneider P. M., Gerke V., Thomas M., Berdel W. E., Serve H., Müller-Tidow C. (2004) Cancer Res. 64, 5564–5569 [DOI] [PubMed] [Google Scholar]
- 6.Yang L., Zhang L., Wu Q., Boyd D. D. (2008) J. Biol. Chem. 283, 35295–35304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szmola R., Kukor Z., Sahin-Tóth M. (2003) J. Biol. Chem. 278, 48580–48589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grishina Z., Ostrowska E., Halangk W., Sahin-Tóth M., Reiser G. (2005) Br. J. Pharmacol. 146, 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knecht W., Cottrell G. S., Amadesi S., Mohlin J., Skåregärde A., Gedda K., Peterson A., Chapman K., Hollenberg M. D., Vergnolle N., Bunnett N. W. (2007) J. Biol. Chem. 282, 26089–26100 [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Luo W., Wartmann T., Halangk W., Sahin-Tóth M., Reiser G. (2006) J. Neurochem. 99, 759–769 [DOI] [PubMed] [Google Scholar]
- 11.Salameh M. A., Soares A. S., Hockla A., Radisky E. S. (2008) J. Biol. Chem. 283, 4115–4123 [DOI] [PubMed] [Google Scholar]
- 12.Bode W., Huber R. (1992) Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 13.Apostoluk W., Otlewski J. (1998) Proteins 32, 459–474 [PubMed] [Google Scholar]
- 14.Krowarsch D., Cierpicki T., Jelen F., Otlewski J. (2003) Cell Mol. Life Sci. 60, 2427–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings N. D., Tolle D. P., Barrett A. J. (2004) Biochem. J. 378, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski M., Jr., Kato I. (1980) Annu. Rev. Biochem. 49, 593–626 [DOI] [PubMed] [Google Scholar]
- 17.Radisky E. S., Koshland D. E., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10316–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) Nucleic Acids Res. 36, D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golde T. E., Estus S., Usiak M., Younkin L. H., Younkin S. G. (1990) Neuron 4, 253–267 [DOI] [PubMed] [Google Scholar]
- 20.Ponte P., Gonzalez-DeWhitt P., Schilling J., Miller J., Hsu D., Greenberg B., Davis K., Wallace W., Lieberburg I., Fuller F. (1988) Nature 331, 525–527 [DOI] [PubMed] [Google Scholar]
- 21.Tanzi R. E., McClatchey A. I., Lamperti E. D., Villa-Komaroff L., Gusella J. F., Neve R. L. (1988) Nature 331, 528–530 [DOI] [PubMed] [Google Scholar]
- 22.Golde T. E., Dickson D., Hutton M. (2006) Curr. Alzheimer Res. 3, 421–430 [DOI] [PubMed] [Google Scholar]
- 23.Smith R. P., Higuchi D. A., Broze G. J., Jr. (1990) Science 248, 1126–1128 [DOI] [PubMed] [Google Scholar]
- 24.Van Nostrand W. E., Wagner S. L., Farrow J. S., Cunningham D. D. (1990) J. Biol. Chem. 265, 9591–9594 [PubMed] [Google Scholar]
- 25.Badellino K. O., Walsh P. N. (2000) Biochemistry 39, 4769–4777 [DOI] [PubMed] [Google Scholar]
- 26.Gralle M., Ferreira S. T. (2007) Prog. Neurobiol. 82, 11–32 [DOI] [PubMed] [Google Scholar]
- 27.Schmitz A., Tikkanen R., Kirfel G., Herzog V. (2002) Histochem. Cell Biol. 117, 171–180 [DOI] [PubMed] [Google Scholar]
- 28.Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. (1987) Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- 29.Jacobsen K. T., Iverfeldt K. (2009) Cell Mol. Life Sci. 66, 2299–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webber M. M., Quader S. T., Kleinman H. K., Bello-DeOcampo D., Storto P. D., Bice G., DeMendonca-Calaca W., Williams D. E. (2001) Prostate 47, 1–13 [DOI] [PubMed] [Google Scholar]
- 31.Dächsel J. C., Taylor J. P., Mok S. S., Ross O. A., Hinkle K. M., Bailey R. M., Hines J. H., Szutu J., Madden B., Petrucelli L., Farrer M. J. (2007) Parkinsonism Relat. Disord. 13, 382–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase T., Jr., Shaw E. (1967) Biochem. Biophys. Res. Commun. 29, 508–514 [DOI] [PubMed] [Google Scholar]
- 33.Navaneetham D., Jin L., Pandey P., Strickler J. E., Babine R. E., Abdel-Meguid S. S., Walsh P. N. (2005) J. Biol. Chem. 280, 36165–36175 [DOI] [PubMed] [Google Scholar]
- 34.DelMar E. G., Largman C., Brodrick J. W., Geokas M. C. (1979) Anal. Biochem. 99, 316–320 [DOI] [PubMed] [Google Scholar]
- 35.Radisky E. S., King D. S., Kwan G., Koshland D. E., Jr. (2003) Biochemistry 42, 6484–6492 [DOI] [PubMed] [Google Scholar]
- 36.Longstaff C., Campbell A. F., Fersht A. R. (1990) Biochemistry 29, 7339–7347 [DOI] [PubMed] [Google Scholar]
- 37.Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 38.Gallagher S. R. (2001) in Current Protocols in Molecular Biology (Ausubel F. M. ed) pp. 10.0.0–10.1.34, John Wiley & Sons, Inc., New York [Google Scholar]
- 39.Perona J. J., Tsu C. A., Craik C. S., Fletterick R. J. (1993) J. Mol. Biol. 230, 919–933 [DOI] [PubMed] [Google Scholar]
- 40.Scheidig A. J., Hynes T. R., Pelletier L. A., Wells J. A., Kossiakoff A. A. (1997) Protein Sci. 6, 1806–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornish-Bowden A. (1995) Fundamentals of Enzyme Kinetics, pp. 105–108, Portland Press, London [Google Scholar]
- 42.Kitaguchi N., Takahashi Y., Oishi K., Shiojiri S., Tokushima Y., Utsunomiya T., Ito H. (1990) Biochim. Biophys. Acta 1038, 105–113 [DOI] [PubMed] [Google Scholar]
- 43.Van Nostrand W. E., Schmaier A. H., Neiditch B. R., Siegel R. S., Raschke W. C., Sisodia S. S., Wagner S. L. (1994) Biochim. Biophys. Acta 1209, 165–170 [DOI] [PubMed] [Google Scholar]
- 44.Madison E. L., Coombs G. S., Corey D. R. (1995) J. Biol. Chem. 270, 7558–7562 [DOI] [PubMed] [Google Scholar]
- 45.Ding L., Coombs G. S., Strandberg L., Navre M., Corey D. R., Madison E. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7627–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahin-Tóth M. (2000) J. Biol. Chem. 275, 22750–22755 [DOI] [PubMed] [Google Scholar]
- 47.Nemoda Z., Sahin-Tóth M. (2005) J. Biol. Chem. 280, 29645–29652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lijnen H. R., Van Hoef B., Collen D. (1984) Eur. J. Biochem. 144, 541–544 [DOI] [PubMed] [Google Scholar]
- 49.Katona G., Berglund G. I., Hajdu J., Gráf L., Szilágyi L. (2002) J. Mol. Biol. 315, 1209–1218 [DOI] [PubMed] [Google Scholar]
- 50.Wright H. T. (1977) Eur. J. Biochem. 73, 567–578 [DOI] [PubMed] [Google Scholar]
- 51.Hubbard S. J., Campbell S. F., Thornton J. M. (1991) J. Mol. Biol. 220, 507–530 [DOI] [PubMed] [Google Scholar]
- 52.Timmer J. C., Zhu W., Pop C., Regan T., Snipas S. J., Eroshkin A. M., Riedl S. J., Salvesen G. S. (2009) Nat. Struct. Mol. Biol. 16, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamba D., Bauer M., Huber R., Fischer S., Rudolph R., Kohnert U., Bode W. (1996) J. Mol. Biol. 258, 117–135 [DOI] [PubMed] [Google Scholar]
- 54.Walsh P. N., Gailani D. (2006) in Hemostasis and Thrombosis: Basic Principles and Clinical Practice, 5th Ed. (Colman R. W., Marder V. J., Clowes A. W., George J. N., Goldhaber S. Z. eds) pp. 221–233, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 55.Nierodzik M. L., Karpatkin S. (2006) Cancer Cell 10, 355–362 [DOI] [PubMed] [Google Scholar]
- 56.Ruf W., Mueller B. M. (2006) Seminars in Thrombosis and Hemostasis 32, Suppl. 1, 61–68 [DOI] [PubMed] [Google Scholar]
- 57.Itoh H., Kataoka H., Koita H., Nabeshima K., Inoue T., Kangawa K., Koono M. (1991) Int. J. Cancer 49, 436–443 [DOI] [PubMed] [Google Scholar]
- 58.Kataoka H., Seguchi K., Iwamura T., Moriyama T., Nabeshima K., Koono M. (1995) Int. J. Cancer 60, 123–128 [DOI] [PubMed] [Google Scholar]
- 59.Seguchi K., Kataoka H., Uchino H., Nabeshima K., Koono M. (1999) Biol. Chem. 380, 473–483 [DOI] [PubMed] [Google Scholar]
- 60.Takayama K., Tsutsumi S., Suzuki T., Horie-Inoue K., Ikeda K., Kaneshiro K., Fujimura T., Kumagai J., Urano T., Sakaki Y., Shirahige K., Sasano H., Takahashi S., Kitamura T., Ouchi Y., Aburatani H., Inoue S. (2009) Cancer Res. 69, 137–142 [DOI] [PubMed] [Google Scholar]
- 61.Kounnas M. Z., Moir R. D., Rebeck G. W., Bush A. I., Argraves W. S., Tanzi R. E., Hyman B. T., Strickland D. K. (1995) Cell 82, 331–340 [DOI] [PubMed] [Google Scholar]
- 62.Németh A. L., Medveczky P., Tóth J., Siklódi E., Schlett K., Patthy A., Palkovits M., Ovádi J., Tõkési N., Németh P., Szilágyi L., Gráf L. (2007) FEBS J. 274, 1610–1620 [DOI] [PubMed] [Google Scholar]
- 63.Tóth J., Siklódi E., Medveczky P., Gallatz K., Németh P., Szilágyi L., Gráf L., Palkovits M. (2007) Neurochem. Res. 32, 1423–1433 [DOI] [PubMed] [Google Scholar]
- 64.Wiegand U., Corbach S., Minn A., Kang J., Müller-Hill B. (1993) Gene 136, 167–175 [DOI] [PubMed] [Google Scholar]
- 65.Xu F., Davis J., Miao J., Previti M. L., Romanov G., Ziegler K., Van Nostrand W. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu F., Previti M. L., Nieman M. T., Davis J., Schmaier A. H., Van Nostrand W. E. (2009) J. Neurosci. 29, 5666–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu F., Previti M. L., Van Nostrand W. E. (2007) Stroke 38, 2598–2601 [DOI] [PubMed] [Google Scholar]
- 68.Qiu W. Q., Ferreira A., Miller C., Koo E. H., Selkoe D. J. (1995) J. Neurosci. 15, 2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gralle M., Oliveira C. L., Guerreiro L. H., McKinstry W. J., Galatis D., Masters C. L., Cappai R., Parker M. W., Ramos C. H., Torriani I., Ferreira S. T. (2006) J. Mol. Biol. 357, 493–508 [DOI] [PubMed] [Google Scholar]