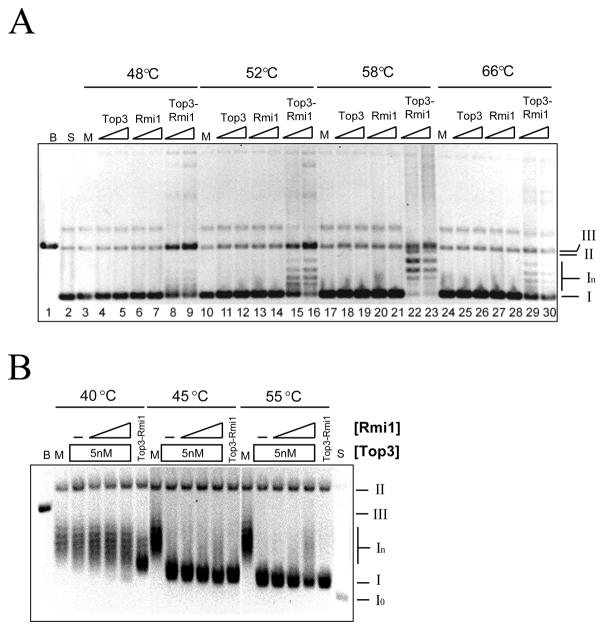
Fig. 2.
Relaxation of negatively supercoiled DNA by Top3, Rmi1, and Top3-Rmi1. (A) Relaxation reactions containing 200 ng pKS+ supercoiled plasmid DNA were incubated at the indicated temperature for 20 min. Following resolution on a native agarose gel, the products were stained with EtBr and photographed. Presented is the negative image. Top3 concentrations were 75 nM or 150 nM; Rmi1 concentrations were 75 nM or 150 nM; and the concentrations of Top3-Rmi1 complex were 45 nM or 90 nM. (B) Relaxation reactions containing 200 ng negatively supercoiled pKS+ plasmid DNA were incubated at the indicated temperature with 5 nM Top3-Rmi1, or 5 nM Top3 together with increasing concentrations of Rmi1. Following the reactions, the products were resolved by 0.8% agarose gel electrophoresis in presence of 5 μg/ml chloroquine and the gel was processed as above. The concentrations of Rmi1 were 25 (lanes 4, 10, 16), 100 (lanes 5, 11, 17), or 200 nM (lanes 6, 12, 18). The various topological states of the plasmid are indicated to the right of the gel as follows: I, negatively supercoiled DNA; In, topoisomers of negatively supercoiled DNA; II, open circle and nicked circle forms; III, linear DNA. B, pKS+ plasmid digested with BamHI (lane 1); M, mock incubation; S, substrate relaxed with DNA topoisomerase I.